Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for CUX1
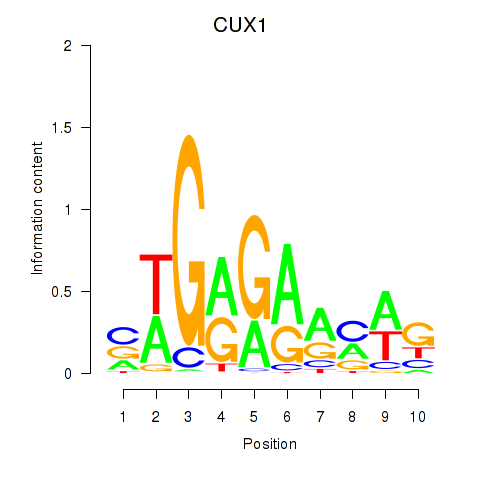
Z-value: 1.26
Transcription factors associated with CUX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX1
|
ENSG00000257923.5 | cut like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX1 | hg19_v2_chr7_+_101917407_101917445 | 0.53 | 3.8e-17 | Click! |
Activity profile of CUX1 motif
Sorted Z-values of CUX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_23241661 | 29.22 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr6_-_32557610 | 28.88 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr17_+_1665345 | 26.72 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr14_-_106092403 | 25.22 |
ENST00000390543.2
|
IGHG4
|
immunoglobulin heavy constant gamma 4 (G4m marker) |
| chr6_-_32498046 | 24.60 |
ENST00000374975.3
|
HLA-DRB5
|
major histocompatibility complex, class II, DR beta 5 |
| chr22_+_23243156 | 21.72 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr2_+_90139056 | 19.29 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr14_+_75745477 | 18.55 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chrX_+_129473859 | 18.54 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr6_-_33048483 | 18.34 |
ENST00000419277.1
|
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chrX_-_153059811 | 17.79 |
ENST00000427365.2
ENST00000444450.1 ENST00000370093.1 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr15_-_22448819 | 17.74 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr5_-_176057518 | 17.68 |
ENST00000393693.2
|
SNCB
|
synuclein, beta |
| chr6_+_31554826 | 17.36 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr5_-_176057365 | 16.66 |
ENST00000310112.3
|
SNCB
|
synuclein, beta |
| chr6_+_31554962 | 16.56 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chrX_-_107019181 | 15.78 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr12_-_91574142 | 15.77 |
ENST00000547937.1
|
DCN
|
decorin |
| chr1_-_183560011 | 15.19 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr11_-_111175739 | 14.94 |
ENST00000532918.1
|
COLCA1
|
colorectal cancer associated 1 |
| chr19_+_41509851 | 14.79 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr12_-_105478339 | 14.66 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr7_-_150038704 | 14.63 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chrX_-_153059958 | 14.30 |
ENST00000370092.3
ENST00000217901.5 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr3_+_49507674 | 14.30 |
ENST00000431960.1
ENST00000452317.1 ENST00000435508.2 ENST00000452060.1 ENST00000428779.1 ENST00000419218.1 ENST00000430636.1 |
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| chr12_+_112204691 | 14.19 |
ENST00000416293.3
ENST00000261733.2 |
ALDH2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr15_+_74908147 | 13.96 |
ENST00000568139.1
ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3
|
CDC-like kinase 3 |
| chr22_-_17680472 | 13.93 |
ENST00000330232.4
|
CECR1
|
cat eye syndrome chromosome region, candidate 1 |
| chr14_-_106322288 | 13.89 |
ENST00000390559.2
|
IGHM
|
immunoglobulin heavy constant mu |
| chr3_+_169491171 | 13.73 |
ENST00000356716.4
|
MYNN
|
myoneurin |
| chr17_+_1665253 | 13.69 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr22_+_23040274 | 13.20 |
ENST00000390306.2
|
IGLV2-23
|
immunoglobulin lambda variable 2-23 |
| chr15_+_75118888 | 13.20 |
ENST00000395018.4
|
CPLX3
|
complexin 3 |
| chr8_-_56685859 | 13.13 |
ENST00000523423.1
ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68
|
transmembrane protein 68 |
| chr1_-_67142710 | 12.73 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr8_-_56685966 | 12.70 |
ENST00000334667.2
|
TMEM68
|
transmembrane protein 68 |
| chr1_+_22962948 | 12.68 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr19_+_5681011 | 12.64 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chrX_-_70331298 | 12.44 |
ENST00000456850.2
ENST00000473378.1 ENST00000487883.1 ENST00000374202.2 |
IL2RG
|
interleukin 2 receptor, gamma |
| chr8_+_27168988 | 12.39 |
ENST00000397501.1
ENST00000338238.4 ENST00000544172.1 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr1_+_11866270 | 12.31 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr1_+_161185032 | 12.25 |
ENST00000367992.3
ENST00000289902.1 |
FCER1G
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr1_+_160085501 | 12.25 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr5_-_131826457 | 12.16 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr9_-_115983641 | 12.07 |
ENST00000238256.3
|
FKBP15
|
FK506 binding protein 15, 133kDa |
| chrX_+_70316005 | 11.91 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr19_+_56652686 | 11.84 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr14_-_21270995 | 11.70 |
ENST00000555698.1
ENST00000397970.4 ENST00000340900.3 |
RNASE1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr1_+_168148273 | 11.68 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr22_+_23077065 | 11.63 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr2_-_96811170 | 11.61 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr12_-_53601000 | 11.59 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr22_+_22786288 | 11.56 |
ENST00000390301.2
|
IGLV1-36
|
immunoglobulin lambda variable 1-36 |
| chr17_-_74137374 | 11.56 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr1_-_112281875 | 11.53 |
ENST00000527621.1
ENST00000534365.1 ENST00000357260.5 |
FAM212B
|
family with sequence similarity 212, member B |
| chr11_-_66725837 | 11.46 |
ENST00000393958.2
ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC
|
pyruvate carboxylase |
| chr1_+_156119798 | 11.39 |
ENST00000355014.2
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_-_27952741 | 11.38 |
ENST00000399173.1
|
FGR
|
feline Gardner-Rasheed sarcoma viral oncogene homolog |
| chr22_-_36903069 | 11.37 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr22_+_23165153 | 11.29 |
ENST00000390317.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr22_-_37545972 | 11.23 |
ENST00000216223.5
|
IL2RB
|
interleukin 2 receptor, beta |
| chr22_+_22712087 | 11.22 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr17_+_41006095 | 11.00 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase, copper containing 3 |
| chr20_-_43883197 | 10.84 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr2_-_89278535 | 10.68 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr14_-_21490417 | 10.63 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chr17_+_47865917 | 10.59 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr7_-_37024665 | 10.59 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr19_-_36523709 | 10.54 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr19_+_56652556 | 10.51 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr5_+_178368186 | 10.45 |
ENST00000320129.3
ENST00000519564.1 |
ZNF454
|
zinc finger protein 454 |
| chr5_-_149535421 | 10.38 |
ENST00000261799.4
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr1_+_89829610 | 10.37 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr1_-_212208842 | 10.36 |
ENST00000366992.3
ENST00000366993.3 ENST00000440600.2 ENST00000366994.3 |
INTS7
|
integrator complex subunit 7 |
| chr3_+_169490606 | 10.35 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr8_-_22089845 | 10.22 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr19_+_45417812 | 10.21 |
ENST00000592535.1
|
APOC1
|
apolipoprotein C-I |
| chr22_-_37213045 | 10.12 |
ENST00000406910.2
ENST00000417718.2 |
PVALB
|
parvalbumin |
| chr17_-_47755338 | 10.08 |
ENST00000508805.1
ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP
|
speckle-type POZ protein |
| chr14_-_21490590 | 10.08 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chrX_+_23352133 | 10.06 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr1_+_46668994 | 10.05 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1 |
| chr21_+_34144411 | 10.05 |
ENST00000382375.4
ENST00000453404.1 ENST00000382378.1 ENST00000477513.1 |
C21orf49
|
chromosome 21 open reading frame 49 |
| chr20_-_18774614 | 10.04 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr12_-_52715179 | 10.02 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr13_-_28674693 | 9.94 |
ENST00000537084.1
ENST00000241453.7 ENST00000380982.4 |
FLT3
|
fms-related tyrosine kinase 3 |
| chr3_-_15106747 | 9.93 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chr8_-_110620284 | 9.93 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr7_+_6522922 | 9.85 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr17_+_7942335 | 9.73 |
ENST00000380183.4
ENST00000572022.1 ENST00000380173.2 |
ALOX15B
|
arachidonate 15-lipoxygenase, type B |
| chr3_+_111717600 | 9.69 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr11_-_118213360 | 9.67 |
ENST00000529594.1
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex) |
| chr4_+_123844225 | 9.64 |
ENST00000274008.4
|
SPATA5
|
spermatogenesis associated 5 |
| chr1_-_203274418 | 9.61 |
ENST00000457348.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr7_-_76829125 | 9.60 |
ENST00000248598.5
|
FGL2
|
fibrinogen-like 2 |
| chr14_-_25045446 | 9.56 |
ENST00000216336.2
|
CTSG
|
cathepsin G |
| chr13_-_50018241 | 9.43 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr12_-_71182695 | 9.39 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr1_+_158801095 | 9.37 |
ENST00000368141.4
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr7_+_65579799 | 9.34 |
ENST00000431089.2
ENST00000398684.2 ENST00000338592.5 |
CRCP
|
CGRP receptor component |
| chr4_-_48018580 | 9.28 |
ENST00000514170.1
|
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr6_+_72926145 | 9.18 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr17_-_2415169 | 9.12 |
ENST00000263092.6
ENST00000538844.1 ENST00000576976.1 |
METTL16
|
methyltransferase like 16 |
| chr1_+_156123359 | 9.12 |
ENST00000368284.1
ENST00000368286.2 ENST00000438830.1 |
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_11866207 | 9.08 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr1_+_160097462 | 9.07 |
ENST00000447527.1
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr19_-_49945617 | 9.01 |
ENST00000600601.1
ENST00000543531.1 |
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr17_+_7452336 | 9.00 |
ENST00000293826.4
|
TNFSF12-TNFSF13
|
TNFSF12-TNFSF13 readthrough |
| chr11_-_72385437 | 8.94 |
ENST00000418754.2
ENST00000542969.2 ENST00000334456.5 |
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr1_+_156123318 | 8.81 |
ENST00000368285.3
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr11_+_118175596 | 8.64 |
ENST00000528600.1
|
CD3E
|
CD3e molecule, epsilon (CD3-TCR complex) |
| chr2_-_73869508 | 8.64 |
ENST00000272425.3
|
NAT8
|
N-acetyltransferase 8 (GCN5-related, putative) |
| chr17_-_18218270 | 8.64 |
ENST00000321105.5
|
TOP3A
|
topoisomerase (DNA) III alpha |
| chr6_+_26204825 | 8.63 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr6_-_34113856 | 8.39 |
ENST00000538487.2
|
GRM4
|
glutamate receptor, metabotropic 4 |
| chr13_-_50018140 | 8.39 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr6_+_31554636 | 8.36 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr3_-_170303845 | 8.26 |
ENST00000231706.5
|
SLC7A14
|
solute carrier family 7, member 14 |
| chr11_+_126225529 | 8.25 |
ENST00000227495.6
ENST00000444328.2 ENST00000356132.4 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr1_-_15911510 | 8.25 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr8_+_11561660 | 8.24 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr11_+_117070037 | 8.23 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr2_+_60983361 | 8.22 |
ENST00000238714.3
|
PAPOLG
|
poly(A) polymerase gamma |
| chr2_+_64751433 | 8.21 |
ENST00000238856.4
ENST00000422803.1 ENST00000238855.7 |
AFTPH
|
aftiphilin |
| chr19_+_55084438 | 8.16 |
ENST00000439534.1
|
LILRA2
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 |
| chr16_-_89768097 | 8.11 |
ENST00000289805.5
ENST00000335360.7 |
SPATA2L
|
spermatogenesis associated 2-like |
| chr12_+_7055631 | 8.09 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_-_152373216 | 8.09 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr19_+_44556158 | 8.07 |
ENST00000434772.3
ENST00000585552.1 |
ZNF223
|
zinc finger protein 223 |
| chr16_+_69166418 | 8.06 |
ENST00000314423.7
ENST00000562237.1 ENST00000567460.1 ENST00000566227.1 ENST00000352319.4 ENST00000563094.1 |
CIRH1A
|
cirrhosis, autosomal recessive 1A (cirhin) |
| chr22_+_41763274 | 8.01 |
ENST00000406644.3
|
TEF
|
thyrotrophic embryonic factor |
| chr14_-_106330458 | 8.01 |
ENST00000461719.1
|
IGHJ4
|
immunoglobulin heavy joining 4 |
| chr1_+_158901329 | 8.00 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr19_-_51530916 | 8.00 |
ENST00000594768.1
|
KLK11
|
kallikrein-related peptidase 11 |
| chr14_-_23285069 | 7.96 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr20_+_18269121 | 7.90 |
ENST00000377671.3
ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133
|
zinc finger protein 133 |
| chr17_-_18950310 | 7.82 |
ENST00000573099.1
|
GRAP
|
GRB2-related adaptor protein |
| chrX_+_46937745 | 7.82 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chr6_-_22297730 | 7.82 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr15_-_64648273 | 7.78 |
ENST00000607537.1
ENST00000303052.7 ENST00000303032.6 |
CSNK1G1
|
casein kinase 1, gamma 1 |
| chr19_-_42133420 | 7.77 |
ENST00000221954.2
ENST00000600925.1 |
CEACAM4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr6_-_46459675 | 7.71 |
ENST00000306764.7
|
RCAN2
|
regulator of calcineurin 2 |
| chr6_-_29600832 | 7.69 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr19_-_5680499 | 7.67 |
ENST00000587589.1
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr14_-_107283278 | 7.59 |
ENST00000390639.2
|
IGHV7-81
|
immunoglobulin heavy variable 7-81 (non-functional) |
| chr19_-_54784353 | 7.57 |
ENST00000391746.1
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chrY_+_16168097 | 7.57 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr19_+_36266433 | 7.57 |
ENST00000314737.5
|
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr1_+_119957554 | 7.56 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr11_-_14993819 | 7.52 |
ENST00000396372.2
ENST00000361010.3 ENST00000359642.3 ENST00000331587.4 |
CALCA
|
calcitonin-related polypeptide alpha |
| chr6_-_31514333 | 7.51 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr1_+_204797749 | 7.49 |
ENST00000367172.4
ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC
|
neurofascin |
| chr7_-_102252038 | 7.49 |
ENST00000461209.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr2_-_50574856 | 7.43 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr6_-_90025011 | 7.42 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr13_-_25496926 | 7.39 |
ENST00000545981.1
ENST00000381884.4 |
CENPJ
|
centromere protein J |
| chr15_+_58702742 | 7.38 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr12_-_11508520 | 7.27 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chrX_+_153059608 | 7.25 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr17_+_73043301 | 7.21 |
ENST00000322444.6
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr19_+_50380682 | 7.20 |
ENST00000221543.5
|
TBC1D17
|
TBC1 domain family, member 17 |
| chr14_-_25103388 | 7.20 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr14_-_106963409 | 7.17 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr14_-_23285011 | 7.15 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_-_167797887 | 7.12 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr1_-_36948879 | 7.11 |
ENST00000373106.1
ENST00000373104.1 ENST00000373103.1 |
CSF3R
|
colony stimulating factor 3 receptor (granulocyte) |
| chr13_+_111365602 | 7.07 |
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1 |
| chr6_-_31514516 | 7.06 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr8_-_110656995 | 7.02 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr14_+_74416989 | 7.02 |
ENST00000334571.2
ENST00000554920.1 |
COQ6
|
coenzyme Q6 monooxygenase |
| chr9_-_115983568 | 7.01 |
ENST00000446284.1
ENST00000414250.1 |
FKBP15
|
FK506 binding protein 15, 133kDa |
| chrX_-_130037198 | 7.01 |
ENST00000370935.1
ENST00000338144.3 ENST00000394363.1 |
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr5_+_35617940 | 6.93 |
ENST00000282469.6
ENST00000509059.1 ENST00000356031.3 ENST00000510777.1 |
SPEF2
|
sperm flagellar 2 |
| chr6_+_31554456 | 6.92 |
ENST00000339530.4
|
LST1
|
leukocyte specific transcript 1 |
| chr9_-_35665165 | 6.92 |
ENST00000343259.3
ENST00000378387.3 |
ARHGEF39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr15_-_88799384 | 6.87 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr15_-_88799661 | 6.86 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr12_+_57522258 | 6.86 |
ENST00000553277.1
ENST00000243077.3 |
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr1_-_212588157 | 6.84 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr2_+_103035102 | 6.84 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr7_-_142176790 | 6.82 |
ENST00000390369.2
|
TRBV7-4
|
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr2_-_152118352 | 6.81 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr9_+_35605274 | 6.80 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr10_+_75545391 | 6.78 |
ENST00000604524.1
ENST00000605216.1 ENST00000398706.2 |
ZSWIM8
|
zinc finger, SWIM-type containing 8 |
| chr6_+_31554612 | 6.75 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chr12_+_7060432 | 6.74 |
ENST00000318974.9
ENST00000456013.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr12_+_79258444 | 6.73 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr10_+_74927875 | 6.68 |
ENST00000242505.6
|
FAM149B1
|
family with sequence similarity 149, member B1 |
| chr2_+_90153696 | 6.68 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr11_-_59950519 | 6.67 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr12_+_79258547 | 6.66 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chrX_-_78622805 | 6.62 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr8_+_38585704 | 6.59 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr5_+_140186647 | 6.55 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr1_-_52870059 | 6.53 |
ENST00000371566.1
|
ORC1
|
origin recognition complex, subunit 1 |
| chrX_+_102469997 | 6.52 |
ENST00000372695.5
ENST00000372691.3 |
BEX4
|
brain expressed, X-linked 4 |
| chr11_-_62783303 | 6.51 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr16_+_23765948 | 6.50 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr1_-_168698433 | 6.50 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr6_-_90024967 | 6.47 |
ENST00000602399.1
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 40.4 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 5.8 | 28.9 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 4.9 | 14.7 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 4.9 | 14.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 4.6 | 32.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 4.6 | 13.7 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 4.5 | 9.0 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 4.5 | 13.4 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 4.3 | 4.3 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 4.3 | 13.0 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 4.3 | 21.3 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 4.3 | 12.8 | GO:0060437 | lung growth(GO:0060437) |
| 4.1 | 12.3 | GO:0001812 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type I hypersensitivity(GO:0001812) positive regulation of type II hypersensitivity(GO:0002894) positive regulation of mast cell cytokine production(GO:0032765) mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 4.1 | 12.2 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 3.9 | 11.6 | GO:0002663 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 3.7 | 11.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 3.7 | 14.8 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 3.4 | 23.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 3.1 | 12.4 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 3.0 | 12.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 3.0 | 9.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 3.0 | 15.0 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 3.0 | 8.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 3.0 | 11.9 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 2.9 | 11.6 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 2.9 | 14.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 2.7 | 8.2 | GO:0003285 | septum secundum development(GO:0003285) |
| 2.6 | 10.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 2.6 | 7.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 2.6 | 10.2 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 2.5 | 7.6 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 2.5 | 5.1 | GO:0001810 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) regulation of type I hypersensitivity(GO:0001810) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) type I hypersensitivity(GO:0016068) |
| 2.5 | 15.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 2.3 | 18.5 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.3 | 6.9 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 2.1 | 10.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 2.1 | 6.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 2.1 | 8.3 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 2.1 | 10.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 2.0 | 15.8 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.9 | 5.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 1.9 | 9.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 1.9 | 5.6 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 1.9 | 7.4 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 1.8 | 14.8 | GO:0070944 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) |
| 1.8 | 9.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 1.7 | 5.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.7 | 17.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.7 | 5.2 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 1.7 | 5.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 1.7 | 5.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.7 | 6.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.7 | 5.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 1.7 | 6.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.6 | 4.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.6 | 9.7 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 1.6 | 4.7 | GO:0021678 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.6 | 4.7 | GO:0045082 | interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 1.5 | 4.6 | GO:0002125 | maternal aggressive behavior(GO:0002125) positive regulation of female receptivity(GO:0045925) |
| 1.5 | 7.6 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 1.5 | 8.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.5 | 7.4 | GO:0061511 | centriole elongation(GO:0061511) |
| 1.5 | 5.8 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.5 | 8.7 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 1.4 | 17.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.4 | 5.6 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.4 | 8.3 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 1.4 | 4.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 1.3 | 8.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 1.3 | 20.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.3 | 3.8 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 1.3 | 6.3 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 1.2 | 88.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 1.2 | 7.2 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.2 | 6.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 1.2 | 10.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.2 | 8.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.2 | 2.3 | GO:1904589 | regulation of protein import(GO:1904589) |
| 1.2 | 9.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.1 | 9.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 1.1 | 14.9 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 1.1 | 7.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.0 | 3.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 1.0 | 2.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 1.0 | 2.9 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.9 | 7.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.9 | 6.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.9 | 3.6 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.9 | 4.5 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.9 | 2.7 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.9 | 12.5 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.9 | 2.7 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.9 | 7.9 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.9 | 3.5 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.9 | 2.6 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.9 | 10.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.8 | 10.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.8 | 1.7 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.8 | 29.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.8 | 0.8 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.8 | 4.0 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.8 | 3.9 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.8 | 2.3 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.8 | 2.3 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.8 | 11.7 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.8 | 3.1 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.8 | 6.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.8 | 5.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.8 | 3.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.8 | 3.8 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.7 | 21.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.7 | 2.9 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.7 | 24.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.7 | 71.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 2.9 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.7 | 2.9 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.7 | 2.8 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.7 | 9.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.7 | 8.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.7 | 2.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.6 | 6.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.6 | 1.9 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.6 | 2.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.6 | 4.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.6 | 57.5 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.6 | 34.3 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.6 | 26.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.6 | 1.8 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.6 | 2.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.6 | 4.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.6 | 1.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.6 | 2.9 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.6 | 20.7 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.6 | 4.6 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.6 | 8.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.6 | 4.0 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.6 | 4.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.6 | 6.2 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.6 | 8.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.6 | 6.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.5 | 3.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.5 | 4.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.5 | 41.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.5 | 1.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.5 | 17.3 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.5 | 8.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.5 | 7.0 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.5 | 5.5 | GO:0071600 | otic vesicle formation(GO:0030916) otic vesicle morphogenesis(GO:0071600) |
| 0.5 | 4.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) protein O-linked fucosylation(GO:0036066) |
| 0.5 | 5.7 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.5 | 5.2 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.5 | 4.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.5 | 1.4 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.5 | 4.6 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.5 | 8.2 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.5 | 1.8 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.5 | 2.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 3.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 3.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.4 | 17.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.4 | 3.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.4 | 11.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.4 | 11.0 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.4 | 0.8 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 3.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 6.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.4 | 2.0 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 1.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.4 | 10.8 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.4 | 2.4 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.4 | 5.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.4 | 2.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.4 | 35.6 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.4 | 1.5 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.4 | 3.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 1.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.4 | 12.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.4 | 6.8 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.4 | 7.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.3 | 9.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 2.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.3 | 4.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.3 | 1.7 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.3 | 8.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 1.8 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.3 | 0.6 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.3 | 11.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 11.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 0.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 1.9 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 | 1.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.3 | 37.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.3 | 19.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.3 | 3.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.3 | 2.1 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.3 | 5.7 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.3 | 10.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.3 | 0.3 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.2 | 3.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 2.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.5 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.2 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 3.3 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.2 | 4.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 4.9 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.2 | 5.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.2 | 4.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.2 | 1.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 8.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.2 | 8.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 2.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 1.2 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 5.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 2.6 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.2 | 2.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 9.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 3.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.2 | 1.5 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 1.3 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.2 | 1.3 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.2 | 3.9 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 5.3 | GO:0048536 | spleen development(GO:0048536) |
| 0.2 | 2.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 3.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 4.3 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.2 | 10.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 1.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 3.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 6.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 4.2 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.2 | 1.3 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.2 | 7.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 1.9 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 3.8 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.2 | 1.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 | 7.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.2 | 3.0 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 6.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 0.6 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 4.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.2 | 7.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 8.3 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 1.3 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 0.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 8.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.1 | 0.9 | GO:0009100 | glycoprotein metabolic process(GO:0009100) |
| 0.1 | 2.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 3.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 4.0 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 1.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 5.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.1 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 2.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.1 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 0.4 | GO:0035932 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 3.3 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 4.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 3.3 | GO:0002449 | lymphocyte mediated immunity(GO:0002449) |
| 0.1 | 1.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 5.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 1.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.1 | 25.8 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.1 | 1.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 3.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 1.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 17.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 2.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 3.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 5.6 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 1.9 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 6.8 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 6.2 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.1 | 2.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 3.1 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.1 | 24.3 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.1 | 7.1 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 1.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 2.3 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.6 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 3.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 8.4 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 2.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 4.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 16.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 4.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 14.0 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.1 | 2.9 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 2.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 10.0 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 2.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.0 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 1.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 2.6 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 1.3 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 1.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 3.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.6 | GO:0002223 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 2.4 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.5 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 3.5 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.2 | GO:0031668 | cellular response to extracellular stimulus(GO:0031668) |
| 0.0 | 2.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.7 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 39.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 3.4 | 71.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 3.4 | 40.4 | GO:0043203 | axon hillock(GO:0043203) |
| 3.0 | 9.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 2.9 | 11.6 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 2.8 | 22.4 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 2.6 | 7.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.5 | 20.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 2.5 | 12.7 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 2.3 | 18.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.9 | 11.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 1.9 | 7.5 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.6 | 81.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.6 | 15.6 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 1.5 | 7.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 1.5 | 4.4 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 1.5 | 2.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.4 | 9.5 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.3 | 8.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.3 | 17.0 | GO:0097433 | dense body(GO:0097433) |
| 1.2 | 25.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 1.2 | 8.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 1.2 | 5.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 1.2 | 10.4 | GO:0032039 | integrator complex(GO:0032039) |
| 1.1 | 15.8 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.1 | 4.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.9 | 10.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.8 | 7.6 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.7 | 2.9 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.7 | 17.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.7 | 10.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 5.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.7 | 9.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.7 | 11.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.6 | 9.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.6 | 10.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.5 | 7.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 5.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 3.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 3.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 4.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.5 | 2.5 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.5 | 6.5 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.5 | 2.9 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.5 | 4.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.5 | 3.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.5 | 4.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 7.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 6.0 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.4 | 9.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 4.5 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.4 | 13.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.4 | 7.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.4 | 5.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 1.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.4 | 34.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.4 | 33.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 11.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 4.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 1.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.3 | 14.6 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.3 | 1.8 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 22.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.3 | 18.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 29.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 1.8 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.3 | 10.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 33.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 8.0 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 5.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.5 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 25.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 39.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 55.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 8.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 4.6 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.2 | 7.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 7.6 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 12.2 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.2 | 3.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 5.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 12.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 0.5 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.2 | 3.7 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 2.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 11.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 40.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 4.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 5.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 2.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 12.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 12.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 4.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 10.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 17.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 6.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 5.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 5.3 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 2.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 53.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.9 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 161.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.5 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 8.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 32.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 5.9 | 23.7 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 4.9 | 14.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 4.1 | 12.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 3.9 | 11.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 3.7 | 11.0 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 3.5 | 10.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 3.2 | 9.6 | GO:0042806 | fucose binding(GO:0042806) |
| 3.0 | 9.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 3.0 | 15.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 3.0 | 12.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 2.8 | 11.4 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 2.7 | 13.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 2.6 | 10.5 | GO:0097001 | ceramide binding(GO:0097001) |
| 2.6 | 7.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 2.6 | 7.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.5 | 7.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 2.5 | 34.3 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 2.4 | 9.7 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 2.4 | 14.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 2.3 | 9.1 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 2.3 | 9.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.2 | 17.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 2.1 | 8.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 2.0 | 20.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 1.9 | 11.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 1.9 | 5.8 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 1.9 | 69.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 1.8 | 7.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.8 | 15.8 | GO:0043426 | MRF binding(GO:0043426) |
| 1.7 | 8.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.7 | 29.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.7 | 6.9 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 1.7 | 6.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 1.7 | 8.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 1.6 | 81.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 1.6 | 12.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 1.6 | 25.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 1.5 | 3.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 1.5 | 8.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 1.5 | 4.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 1.4 | 8.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.4 | 4.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.4 | 4.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 1.4 | 15.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 1.3 | 10.7 | GO:0032396 | inhibitory MHC class I receptor activity(GO:0032396) |
| 1.3 | 9.3 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 1.3 | 3.9 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.3 | 14.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 1.3 | 7.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.3 | 10.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.2 | 15.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 1.2 | 4.9 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.2 | 3.5 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.1 | 11.2 | GO:0009374 | biotin binding(GO:0009374) |
| 1.1 | 13.7 | GO:0005030 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 1.0 | 8.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 1.0 | 8.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.0 | 21.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.9 | 17.4 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.9 | 9.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.9 | 3.5 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.8 | 3.4 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.8 | 3.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.8 | 5.6 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.8 | 3.2 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.8 | 3.1 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.8 | 7.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.7 | 8.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.7 | 4.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 8.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.7 | 10.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.7 | 2.9 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.7 | 12.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 9.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.7 | 5.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.7 | 18.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 9.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.7 | 4.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.7 | 6.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 10.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 18.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.7 | 2.7 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.7 | 5.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.7 | 3.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.6 | 4.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.6 | 89.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.6 | 3.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 10.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.6 | 6.1 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.6 | 5.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.6 | 4.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.6 | 2.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.5 | 9.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.5 | 4.9 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.5 | 2.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 6.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.5 | 6.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.5 | 18.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.5 | 13.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.5 | 2.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.5 | 2.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.5 | 10.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.5 | 7.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 52.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 24.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.5 | 3.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.5 | 2.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 13.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.5 | 5.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 2.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.4 | 8.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.4 | 8.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.4 | 3.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.4 | 5.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 2.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.4 | 11.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.4 | 5.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 6.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.4 | 1.9 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.4 | 4.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 1.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 4.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.4 | 2.9 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.4 | 13.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 11.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 2.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.3 | 2.7 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.3 | 5.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 3.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 2.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.8 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.3 | 7.9 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.3 | 7.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 5.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.3 | 20.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.3 | 7.0 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 3.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 0.8 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 1.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.3 | 7.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 2.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.8 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 1.3 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 5.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 11.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 53.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.2 | 1.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 4.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 2.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 3.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 15.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 12.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 2.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 4.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 7.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 3.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 8.4 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.2 | 1.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 6.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 9.3 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.2 | 1.7 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 1.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.2 | 2.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 4.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 2.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 9.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 6.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 3.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 11.4 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.2 | 3.1 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.2 | 7.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.8 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 7.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.2 | 5.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 1.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 8.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 2.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.4 | GO:0032027 | fibronectin binding(GO:0001968) myosin light chain binding(GO:0032027) |
| 0.2 | 11.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 10.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 1.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 6.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 3.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 3.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 5.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 10.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 4.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.1 | 2.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 7.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 5.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 14.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 2.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 7.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 14.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 2.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 9.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 9.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 33.7 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 95.7 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 1.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.1 | 0.3 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 2.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.2 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 1.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 3.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 11.1 | GO:0016746 | transferase activity, transferring acyl groups(GO:0016746) |
| 0.1 | 4.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 4.8 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 14.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.6 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.1 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 86.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 1.6 | 17.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.7 | 13.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 26.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.6 | 16.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 13.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 4.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.5 | 26.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.4 | 16.8 | PID EPO PATHWAY | EPO signaling pathway |
| 0.4 | 14.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.4 | 5.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 8.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.4 | 8.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.4 | 21.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 10.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.3 | 3.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 16.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 7.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 46.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 19.7 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 7.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 7.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 13.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 7.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 3.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 12.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 6.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 51.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 4.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 23.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 9.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 8.6 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 4.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 3.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 6.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 11.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 10.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.2 | 6.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 7.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 9.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 2.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 4.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 96.5 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 2.2 | 17.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 1.3 | 32.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.3 | 34.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.2 | 29.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 1.0 | 9.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 1.0 | 20.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.9 | 18.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.9 | 16.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.9 | 16.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.8 | 12.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 17.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.7 | 32.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.7 | 10.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.7 | 10.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.7 | 7.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 9.0 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.6 | 16.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 9.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.6 | 12.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.6 | 6.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.6 | 8.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.6 | 22.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 11.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.6 | 4.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.6 | 23.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.5 | 6.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.5 | 3.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 13.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.5 | 23.6 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.5 | 25.0 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.5 | 8.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.5 | 9.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 4.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 2.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.4 | 15.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.4 | 4.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 5.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.4 | 8.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 31.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.4 | 14.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 7.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 5.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 29.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 10.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 3.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 4.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 19.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.3 | 10.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.3 | 11.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.3 | 14.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 5.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 6.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 4.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 5.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 13.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 3.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 2.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 2.7 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 1.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 5.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 2.9 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 1.4 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 3.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 6.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.5 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 6.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 10.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 2.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 3.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 9.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 2.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 4.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 6.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.7 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |