Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
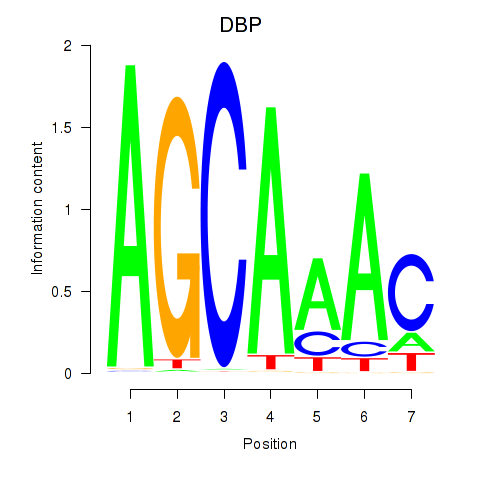
Results for DBP
Z-value: 0.11
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49140609_49140643 | 0.19 | 4.8e-03 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_57037284 | 14.42 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr4_-_176733897 | 11.11 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr20_+_11898507 | 8.76 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr11_+_121461097 | 8.57 |
ENST00000527934.1
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr4_+_158142750 | 7.07 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr8_+_85618155 | 6.62 |
ENST00000523850.1
ENST00000521376.1 |
RALYL
|
RALY RNA binding protein-like |
| chr5_-_111091948 | 6.55 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr3_-_33700589 | 6.10 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_54755459 | 5.96 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr7_-_150946015 | 5.56 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr8_-_102803163 | 5.55 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr1_+_84609944 | 5.43 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr6_-_31514516 | 5.26 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr6_-_31514333 | 5.18 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr16_+_7382745 | 5.06 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr4_+_113970772 | 4.70 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr15_+_80733570 | 4.60 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chrX_-_10851762 | 4.54 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr18_-_21891460 | 4.40 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr11_-_122931881 | 4.40 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr3_+_123813509 | 4.38 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chrX_+_56590002 | 4.36 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr7_+_26331541 | 4.21 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr13_-_67802549 | 4.07 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr7_-_37026108 | 3.92 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_-_80993010 | 3.92 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr4_+_71587669 | 3.83 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_+_87797351 | 3.78 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr14_+_29234870 | 3.67 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr2_+_210517895 | 3.58 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr9_-_139371533 | 3.58 |
ENST00000290037.6
ENST00000431893.2 ENST00000371706.3 |
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr11_+_35684288 | 3.41 |
ENST00000299413.5
|
TRIM44
|
tripartite motif containing 44 |
| chr9_-_139372141 | 3.35 |
ENST00000313050.7
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr8_+_85095553 | 3.30 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr8_+_85095769 | 3.17 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr3_-_33700933 | 3.11 |
ENST00000480013.1
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_210444748 | 3.08 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr12_-_89746173 | 3.06 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr4_-_74088800 | 3.02 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr5_-_134735568 | 2.92 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr4_-_186732048 | 2.90 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_60780536 | 2.90 |
ENST00000538214.1
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr4_-_87281224 | 2.90 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_114214125 | 2.86 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr4_-_87281196 | 2.71 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_+_107738240 | 2.71 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr2_-_88427568 | 2.66 |
ENST00000393750.3
ENST00000295834.3 |
FABP1
|
fatty acid binding protein 1, liver |
| chr8_+_9953061 | 2.57 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr8_+_9953214 | 2.56 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chrX_+_23801280 | 2.52 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr8_-_120685608 | 2.45 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_-_60780702 | 2.42 |
ENST00000359629.5
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr11_-_31832581 | 2.40 |
ENST00000379111.2
|
PAX6
|
paired box 6 |
| chr6_-_75953484 | 2.32 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr17_+_2240775 | 2.26 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr3_+_35721106 | 2.21 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr20_-_14318248 | 2.16 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr1_-_143767881 | 2.09 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr2_+_210444142 | 2.05 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr6_+_21593972 | 2.01 |
ENST00000244745.1
ENST00000543472.1 |
SOX4
|
SRY (sex determining region Y)-box 4 |
| chr4_-_57547454 | 2.00 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr22_+_38201114 | 2.00 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr14_+_29236269 | 1.93 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr8_-_22550815 | 1.91 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr8_-_17533838 | 1.87 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr4_-_16900410 | 1.85 |
ENST00000304523.5
|
LDB2
|
LIM domain binding 2 |
| chr6_+_30687978 | 1.82 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr17_+_7487146 | 1.82 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr6_-_56819385 | 1.80 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr4_-_16900242 | 1.80 |
ENST00000502640.1
ENST00000506732.1 |
LDB2
|
LIM domain binding 2 |
| chr17_+_2240916 | 1.76 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr2_-_190044480 | 1.74 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr4_-_16900184 | 1.71 |
ENST00000515064.1
|
LDB2
|
LIM domain binding 2 |
| chr6_+_10528560 | 1.68 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr19_-_42806919 | 1.65 |
ENST00000595530.1
ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr2_-_65357225 | 1.64 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr4_-_16900217 | 1.61 |
ENST00000441778.2
|
LDB2
|
LIM domain binding 2 |
| chr8_+_26247878 | 1.61 |
ENST00000518611.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr11_+_64009072 | 1.59 |
ENST00000535135.1
ENST00000394540.3 |
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr2_+_166326157 | 1.57 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr3_-_48471454 | 1.53 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr5_+_161495038 | 1.50 |
ENST00000393933.4
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_+_229440129 | 1.50 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr20_+_60718785 | 1.48 |
ENST00000421564.1
ENST00000450482.1 ENST00000331758.3 |
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr9_+_2158485 | 1.44 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr4_+_71588372 | 1.44 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr12_-_120765565 | 1.42 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr6_+_3118926 | 1.41 |
ENST00000380379.5
|
BPHL
|
biphenyl hydrolase-like (serine hydrolase) |
| chr19_-_48894104 | 1.41 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr18_+_32621324 | 1.37 |
ENST00000300249.5
ENST00000538170.2 ENST00000588910.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_+_190333097 | 1.37 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr13_+_97874574 | 1.32 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_+_112832090 | 1.30 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr4_+_160188889 | 1.27 |
ENST00000264431.4
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr5_+_161494770 | 1.27 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr5_+_161494521 | 1.23 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr2_+_234602305 | 1.22 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_-_21266935 | 1.21 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr17_+_72426891 | 1.19 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr9_+_2158443 | 1.19 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_+_42429493 | 1.17 |
ENST00000586242.1
|
GRN
|
granulin |
| chr3_-_185538849 | 1.16 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_+_154229547 | 1.15 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr17_+_48823975 | 1.14 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_+_155051305 | 1.14 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr3_+_109128837 | 1.11 |
ENST00000497996.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr15_+_58702742 | 1.07 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr8_+_104310661 | 1.06 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr3_-_145878954 | 1.06 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr1_+_206557366 | 1.00 |
ENST00000414007.1
ENST00000419187.2 |
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr17_+_4613918 | 0.98 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr7_-_104909435 | 0.96 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr7_+_99613212 | 0.95 |
ENST00000426572.1
ENST00000535170.1 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr10_+_47894572 | 0.95 |
ENST00000355876.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr4_-_89978299 | 0.94 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr5_+_179921344 | 0.89 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr19_+_6361440 | 0.84 |
ENST00000245816.4
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr11_+_112832202 | 0.82 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr17_+_79953310 | 0.81 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr12_-_56236734 | 0.80 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr20_+_36405665 | 0.78 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr1_+_27648709 | 0.78 |
ENST00000608611.1
ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222
|
transmembrane protein 222 |
| chr20_+_361261 | 0.77 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chrX_+_129473916 | 0.74 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr17_+_4613776 | 0.71 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr17_+_48624450 | 0.69 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr4_+_39699664 | 0.69 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chr15_+_78632666 | 0.68 |
ENST00000299529.6
|
CRABP1
|
cellular retinoic acid binding protein 1 |
| chr3_+_155838337 | 0.67 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr11_-_62414070 | 0.67 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr1_+_154540246 | 0.66 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chr15_-_63450192 | 0.66 |
ENST00000411926.1
|
RPS27L
|
ribosomal protein S27-like |
| chr6_+_44194762 | 0.65 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr5_+_179921430 | 0.63 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr1_-_23886285 | 0.62 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr5_+_140710061 | 0.61 |
ENST00000517417.1
ENST00000378105.3 |
PCDHGA1
|
protocadherin gamma subfamily A, 1 |
| chr7_-_22234381 | 0.61 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr16_+_29789561 | 0.60 |
ENST00000400752.4
|
ZG16
|
zymogen granule protein 16 |
| chr8_-_29120580 | 0.59 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr10_-_104178857 | 0.58 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr16_+_19183671 | 0.58 |
ENST00000562711.2
|
SYT17
|
synaptotagmin XVII |
| chr18_-_25616519 | 0.56 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr4_+_39699776 | 0.56 |
ENST00000503368.1
ENST00000445950.2 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chr7_-_83824169 | 0.54 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_155107820 | 0.47 |
ENST00000484157.1
|
SLC50A1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr14_-_23652849 | 0.46 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chrX_+_49028265 | 0.46 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr3_-_47950745 | 0.45 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr6_-_86352982 | 0.45 |
ENST00000369622.3
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr2_-_152830479 | 0.44 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_+_140588269 | 0.43 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr17_+_67498538 | 0.42 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr6_-_33168391 | 0.41 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr16_+_30960375 | 0.40 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr5_+_72143988 | 0.40 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr8_+_134125727 | 0.39 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr7_+_148892557 | 0.38 |
ENST00000262085.3
|
ZNF282
|
zinc finger protein 282 |
| chr16_-_18468926 | 0.37 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_31582961 | 0.35 |
ENST00000376059.3
ENST00000337917.7 |
AIF1
|
allograft inflammatory factor 1 |
| chr14_+_24590560 | 0.35 |
ENST00000558325.1
|
RP11-468E2.6
|
RP11-468E2.6 |
| chr19_+_24009879 | 0.35 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr1_+_104293028 | 0.34 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr15_+_71228826 | 0.33 |
ENST00000558456.1
ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr20_+_43935474 | 0.30 |
ENST00000372743.1
ENST00000372741.3 ENST00000343694.3 |
RBPJL
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr6_+_26251835 | 0.30 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr10_-_65028817 | 0.28 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr10_+_103892787 | 0.26 |
ENST00000278070.2
ENST00000413464.2 |
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr1_+_104159999 | 0.26 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr3_-_88108212 | 0.21 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr7_-_50633078 | 0.21 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr1_+_28764653 | 0.20 |
ENST00000373836.3
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr6_+_33168637 | 0.20 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_48546232 | 0.20 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chrX_-_124097620 | 0.20 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr5_+_179233376 | 0.19 |
ENST00000376929.3
ENST00000514093.1 |
SQSTM1
|
sequestosome 1 |
| chr10_-_64576105 | 0.19 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr3_+_69812701 | 0.18 |
ENST00000472437.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr17_-_56492989 | 0.18 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr4_-_23891693 | 0.17 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr2_-_174830430 | 0.16 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr1_-_151804314 | 0.15 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr2_+_168725458 | 0.14 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr2_-_136743169 | 0.13 |
ENST00000264161.4
|
DARS
|
aspartyl-tRNA synthetase |
| chr4_+_106473768 | 0.13 |
ENST00000265154.2
ENST00000420470.2 |
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr1_+_57320437 | 0.13 |
ENST00000361249.3
|
C8A
|
complement component 8, alpha polypeptide |
| chrX_+_107288197 | 0.09 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_33168597 | 0.09 |
ENST00000374675.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_48546324 | 0.08 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chrX_+_142967173 | 0.07 |
ENST00000370494.1
|
UBE2NL
|
ubiquitin-conjugating enzyme E2N-like |
| chr6_+_26045603 | 0.06 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr5_-_142780280 | 0.06 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_+_28705921 | 0.06 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr2_+_234601512 | 0.06 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr1_+_153700518 | 0.04 |
ENST00000318967.2
ENST00000456435.1 ENST00000435409.2 |
INTS3
|
integrator complex subunit 3 |
| chr9_+_100745615 | 0.04 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr9_-_131038214 | 0.04 |
ENST00000609374.1
|
GOLGA2
|
golgin A2 |
| chr3_-_149375783 | 0.03 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr4_-_157892498 | 0.03 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr5_+_180682720 | 0.03 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr21_+_34398153 | 0.02 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.6 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 2.5 | 7.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.8 | 14.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.7 | 5.2 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 1.2 | 4.8 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.2 | 9.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 1.1 | 5.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.1 | 4.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.1 | 5.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.0 | 2.9 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.9 | 4.4 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.8 | 2.5 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.8 | 3.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.7 | 5.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 3.4 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.6 | 5.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.6 | 0.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.5 | 1.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.5 | 2.0 | GO:0035910 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.5 | 16.4 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.5 | 2.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.4 | 1.3 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.4 | 1.4 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.4 | 1.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 4.5 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.3 | 3.8 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.3 | 1.7 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.3 | 11.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 1.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.3 | 2.7 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 1.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 1.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 6.8 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 1.7 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.9 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.2 | 3.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 1.4 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.7 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 1.8 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.2 | 1.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 1.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 1.2 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.2 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 3.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.2 | 3.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.7 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) synaptic transmission involved in micturition(GO:0060084) |
| 0.2 | 5.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.2 | 4.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 1.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 0.5 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.1 | 6.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 8.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 6.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.0 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.1 | 0.5 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 1.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 4.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 1.1 | GO:1902959 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) regulation of aspartic-type peptidase activity(GO:1905245) |
| 0.1 | 2.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 0.7 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 1.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 3.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 2.5 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 3.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 4.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.2 | GO:0052314 | serotonin biosynthetic process(GO:0042427) phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.3 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 2.0 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 8.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 4.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.8 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 6.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.7 | GO:1902260 | diaphragm development(GO:0060539) negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 2.4 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 1.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.7 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 2.0 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.1 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.8 | GO:0034605 | cellular response to heat(GO:0034605) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 14.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 1.5 | 8.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.4 | 7.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 1.2 | 6.0 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.9 | 9.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.7 | 4.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.7 | 10.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.6 | 4.4 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.6 | 13.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.6 | 1.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 10.4 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.5 | 2.9 | GO:0001740 | Barr body(GO:0001740) |
| 0.5 | 1.8 | GO:0031673 | H zone(GO:0031673) |
| 0.4 | 9.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.4 | 5.5 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 2.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.3 | 3.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 5.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.8 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.2 | 1.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 7.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 7.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 3.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 1.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 4.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 4.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 2.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 4.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 6.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 3.3 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 8.6 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 5.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 5.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 9.1 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 2.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.2 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 8.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.0 | GO:0070876 | SOSS complex(GO:0070876) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.0 | 5.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.9 | 5.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.7 | 17.9 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.6 | 7.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.6 | 2.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.6 | 1.7 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.5 | 6.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.5 | 8.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 4.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.4 | 2.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 1.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.4 | 1.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.3 | 5.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 4.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 2.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.3 | 1.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.3 | 1.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 1.7 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 6.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 1.4 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 3.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 4.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 2.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.2 | 3.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 5.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 7.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 4.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 3.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 1.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 2.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 10.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 3.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 9.3 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.1 | 4.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 2.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.7 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 4.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.6 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 9.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.9 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 4.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 3.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.5 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 2.1 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 14.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 5.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 7.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 1.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 4.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 7.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 14.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.6 | 16.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 5.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 6.5 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.3 | 3.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 5.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 5.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 5.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 4.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 0.8 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 4.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 3.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |