Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
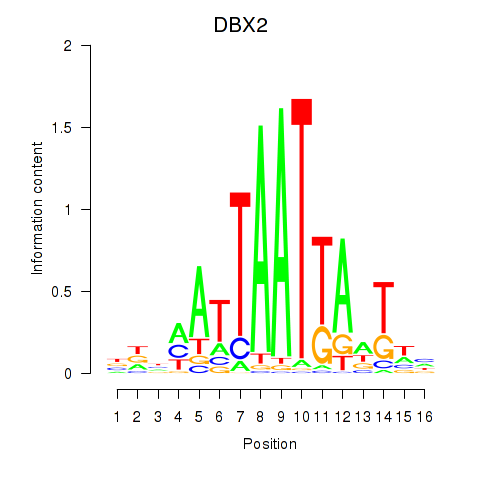
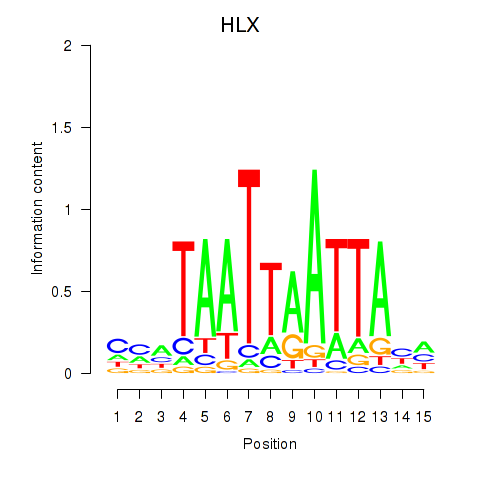
Results for DBX2_HLX
Z-value: 0.51
Transcription factors associated with DBX2_HLX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBX2
|
ENSG00000185610.6 | developing brain homeobox 2 |
|
HLX
|
ENSG00000136630.11 | H2.0 like homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLX | hg19_v2_chr1_+_221051699_221051699 | 0.06 | 4.0e-01 | Click! |
Activity profile of DBX2_HLX motif
Sorted Z-values of DBX2_HLX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_138340049 | 21.20 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_+_113568207 | 20.78 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr3_+_138340067 | 13.30 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr8_-_27695552 | 12.16 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr1_+_81771806 | 11.45 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr1_-_89357179 | 11.31 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr8_+_98900132 | 10.62 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr5_-_146781153 | 10.45 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr11_+_35201826 | 10.27 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_-_45988542 | 9.98 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr2_-_10587897 | 9.36 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr7_-_87856280 | 9.30 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr12_-_10978957 | 9.24 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr3_+_33155525 | 9.14 |
ENST00000449224.1
|
CRTAP
|
cartilage associated protein |
| chr7_-_87856303 | 9.12 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr2_+_187371440 | 8.77 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr2_+_109204909 | 8.38 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chrX_-_77225135 | 8.35 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr17_+_45728427 | 7.94 |
ENST00000540627.1
|
KPNB1
|
karyopherin (importin) beta 1 |
| chr6_+_26104104 | 7.90 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr16_+_15489603 | 7.57 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chrX_+_56590002 | 7.44 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr1_+_224544572 | 6.99 |
ENST00000366857.5
ENST00000366856.3 |
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr3_-_185538849 | 6.99 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_+_102413726 | 6.98 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr2_+_87135076 | 6.62 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr6_+_34204642 | 6.60 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr17_-_8113542 | 6.53 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr14_+_53173910 | 6.53 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr3_-_141719195 | 6.50 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_-_64023441 | 6.48 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr2_-_17981462 | 6.40 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr6_+_34725263 | 6.38 |
ENST00000374018.1
ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr14_+_53173890 | 6.33 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr3_-_141747950 | 6.32 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr8_-_13134045 | 6.22 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr15_+_64680003 | 6.20 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr2_+_201936707 | 6.07 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr22_+_42017987 | 5.86 |
ENST00000405506.1
|
XRCC6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr12_+_104337515 | 5.82 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr7_-_5998714 | 5.78 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr17_+_35851570 | 5.78 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr16_+_24549014 | 5.76 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr2_+_109204743 | 5.66 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr6_-_8102279 | 5.55 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr6_+_89674246 | 5.52 |
ENST00000369474.1
|
AL079342.1
|
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
| chr1_-_110950564 | 5.46 |
ENST00000256644.4
|
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr2_+_161993412 | 5.26 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr7_+_130126165 | 5.16 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr1_-_54411255 | 5.02 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_+_64798095 | 4.94 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chrX_-_16887963 | 4.87 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr4_-_103746683 | 4.83 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_18548426 | 4.82 |
ENST00000357193.3
ENST00000536719.1 |
TSG101
|
tumor susceptibility 101 |
| chr22_-_29107919 | 4.80 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr8_+_132952112 | 4.79 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chrX_-_106243451 | 4.77 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr12_+_20963632 | 4.76 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr10_-_95242044 | 4.73 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr5_+_115177178 | 4.66 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr8_-_101719159 | 4.65 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr15_+_65843130 | 4.65 |
ENST00000569894.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr1_-_212965104 | 4.63 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr12_+_20963647 | 4.60 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr11_-_107729887 | 4.60 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr10_-_95241951 | 4.59 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr5_+_82767284 | 4.58 |
ENST00000265077.3
|
VCAN
|
versican |
| chr9_+_12693336 | 4.53 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr20_-_33735070 | 4.49 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr1_-_153643442 | 4.40 |
ENST00000368681.1
ENST00000361891.4 |
ILF2
|
interleukin enhancer binding factor 2 |
| chr12_+_75874460 | 4.27 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr5_+_82767487 | 4.27 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr7_+_107224364 | 4.20 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr3_+_160939050 | 4.19 |
ENST00000493066.1
ENST00000351193.2 ENST00000472947.1 ENST00000463518.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr16_-_28634874 | 4.14 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chrX_-_135962876 | 4.04 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr7_+_130126012 | 4.03 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr10_+_62538089 | 3.98 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr12_+_123237321 | 3.97 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr2_+_109237717 | 3.90 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr1_+_84630645 | 3.88 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr17_-_60142609 | 3.81 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr4_+_41540160 | 3.78 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr14_-_78083112 | 3.77 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr2_-_113594279 | 3.76 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr11_-_63376013 | 3.73 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr11_-_10828892 | 3.69 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr9_+_125132803 | 3.68 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_+_8191815 | 3.63 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr8_+_31497271 | 3.60 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr1_+_76251879 | 3.56 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr3_+_136649311 | 3.55 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr21_-_18985230 | 3.55 |
ENST00000457956.1
ENST00000348354.6 |
BTG3
|
BTG family, member 3 |
| chr15_+_67418047 | 3.54 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr2_+_109223595 | 3.53 |
ENST00000410093.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr3_+_139063372 | 3.53 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr3_-_123512688 | 3.53 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr14_-_35591433 | 3.50 |
ENST00000261475.5
ENST00000555644.1 |
PPP2R3C
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr9_-_70465758 | 3.47 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr9_-_2844058 | 3.47 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr11_+_101983176 | 3.45 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr5_-_68665084 | 3.45 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr8_-_101718991 | 3.42 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr11_-_104827425 | 3.40 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr20_-_1447467 | 3.39 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr12_-_7656357 | 3.39 |
ENST00000396620.3
ENST00000432237.2 ENST00000359156.4 |
CD163
|
CD163 molecule |
| chr2_-_55496174 | 3.33 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr20_-_1447547 | 3.33 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr7_+_134576317 | 3.32 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chrX_-_114252193 | 3.29 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr2_-_145277569 | 3.24 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_-_102942961 | 3.15 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr6_+_114178512 | 3.14 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr11_-_59950486 | 3.12 |
ENST00000426738.2
ENST00000533023.1 ENST00000420732.2 |
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr8_-_141774467 | 3.11 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr14_+_35591928 | 3.09 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr7_+_116654935 | 2.97 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr20_+_1099233 | 2.96 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr16_+_14802801 | 2.95 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr12_+_75874580 | 2.94 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr20_+_11871433 | 2.94 |
ENST00000399006.2
ENST00000405977.1 |
BTBD3
|
BTB (POZ) domain containing 3 |
| chr20_+_11871371 | 2.94 |
ENST00000254977.3
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr6_+_153552455 | 2.93 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr19_-_13044494 | 2.92 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr3_+_158787041 | 2.88 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr1_-_110283138 | 2.87 |
ENST00000256594.3
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr7_-_140714739 | 2.86 |
ENST00000467334.1
ENST00000324787.5 |
MRPS33
|
mitochondrial ribosomal protein S33 |
| chr19_-_43382142 | 2.83 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr7_+_134576151 | 2.79 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr2_-_58468437 | 2.77 |
ENST00000403676.1
ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL
|
Fanconi anemia, complementation group L |
| chr11_-_327537 | 2.76 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr12_+_75874984 | 2.75 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr7_-_139763521 | 2.75 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chrX_-_100662881 | 2.73 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chrX_-_102943022 | 2.73 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chr9_-_21305312 | 2.69 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr2_-_88285309 | 2.67 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr5_+_95066823 | 2.65 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr1_-_145826450 | 2.59 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr2_-_191115229 | 2.58 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr7_+_129932974 | 2.58 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr2_-_37544209 | 2.57 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr5_-_10761206 | 2.56 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr8_-_49834299 | 2.55 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr16_-_28222797 | 2.54 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr10_+_79793518 | 2.49 |
ENST00000440692.1
ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24
|
ribosomal protein S24 |
| chr2_+_6111712 | 2.48 |
ENST00000391666.2
|
FLJ30594
|
HCG1990367; Putative uncharacterized protein DKFZp761K2322; Putative uncharacterized protein FLJ30594; Uncharacterized protein; cDNA FLJ30594 fis, clone BRAWH2008903 |
| chr5_+_67588391 | 2.47 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_-_49833978 | 2.47 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr3_+_157827841 | 2.44 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr6_+_34725181 | 2.42 |
ENST00000244520.5
|
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr17_-_55038375 | 2.39 |
ENST00000240316.4
|
COIL
|
coilin |
| chr12_+_10658201 | 2.36 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr5_-_139726181 | 2.35 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr2_+_103089756 | 2.35 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr4_-_164534657 | 2.32 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr2_+_86669118 | 2.30 |
ENST00000427678.1
ENST00000542128.1 |
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr5_-_34916871 | 2.29 |
ENST00000382038.2
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr11_+_110300607 | 2.29 |
ENST00000260270.2
|
FDX1
|
ferredoxin 1 |
| chr7_-_22862406 | 2.29 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr16_-_29934558 | 2.27 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr10_-_112678692 | 2.27 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr15_-_42749711 | 2.26 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr10_-_112678904 | 2.26 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr14_+_39583427 | 2.25 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr14_+_89060749 | 2.24 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr1_-_197036364 | 2.22 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr2_+_169312350 | 2.21 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr8_+_11666649 | 2.19 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr10_+_122610687 | 2.16 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr12_+_10658489 | 2.15 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr14_+_89060739 | 2.14 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr10_-_94257512 | 2.05 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr11_-_59950519 | 2.05 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr2_+_11674213 | 2.05 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_-_75960024 | 2.01 |
ENST00000370081.2
|
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr6_+_158733692 | 2.00 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr11_-_104817919 | 1.96 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr11_+_2405833 | 1.95 |
ENST00000527343.1
ENST00000464784.2 |
CD81
|
CD81 molecule |
| chr2_-_37068530 | 1.95 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr4_+_41937131 | 1.94 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr8_-_54752406 | 1.94 |
ENST00000520188.1
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr9_-_21217310 | 1.93 |
ENST00000380216.1
|
IFNA16
|
interferon, alpha 16 |
| chr11_+_44117099 | 1.92 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr4_+_147096837 | 1.92 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_-_11685345 | 1.90 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr4_+_69962212 | 1.87 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr20_-_7921090 | 1.86 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr1_-_35450897 | 1.86 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chrX_+_100663243 | 1.85 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr19_+_54466179 | 1.84 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr1_-_21377383 | 1.83 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr4_-_48782259 | 1.82 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr4_+_69962185 | 1.82 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chrX_-_119445306 | 1.82 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr2_-_70520539 | 1.80 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr17_+_67498538 | 1.80 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_+_45825616 | 1.77 |
ENST00000442528.2
ENST00000456334.1 ENST00000526817.1 |
SLC35C1
|
solute carrier family 35 (GDP-fucose transporter), member C1 |
| chr5_-_125930929 | 1.76 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr10_+_51572339 | 1.75 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBX2_HLX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 2.6 | 18.4 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 2.3 | 9.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 2.2 | 6.5 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 2.1 | 8.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 2.0 | 7.9 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 1.9 | 7.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 1.9 | 11.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 1.9 | 3.8 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.9 | 7.4 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 1.7 | 1.7 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.7 | 5.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 1.7 | 6.6 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 1.6 | 4.8 | GO:2000397 | ubiquitin-dependent endocytosis(GO:0070086) regulation of viral budding via host ESCRT complex(GO:1903772) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 1.6 | 4.8 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.5 | 4.5 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 1.4 | 4.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 1.3 | 4.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.3 | 10.3 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.2 | 3.5 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 1.1 | 6.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 1.0 | 8.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.0 | 5.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 1.0 | 4.9 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.9 | 3.8 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.9 | 3.6 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.9 | 8.1 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.9 | 7.8 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.8 | 9.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.8 | 1.6 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.8 | 3.9 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.8 | 2.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.8 | 4.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.7 | 2.9 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.7 | 3.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.7 | 2.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.7 | 4.0 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.6 | 1.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.6 | 13.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.6 | 1.9 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.6 | 2.6 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.6 | 6.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.6 | 1.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.6 | 1.8 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.6 | 5.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.6 | 1.8 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.6 | 2.9 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.6 | 2.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.6 | 3.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.5 | 3.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 1.6 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.5 | 2.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.5 | 3.6 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.5 | 12.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.5 | 1.9 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.5 | 17.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.5 | 6.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 3.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 2.2 | GO:0097384 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) cellular lipid biosynthetic process(GO:0097384) |
| 0.4 | 3.5 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.4 | 1.7 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.4 | 1.3 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.4 | 4.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 5.8 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.4 | 6.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 1.7 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.4 | 10.8 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.4 | 5.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.4 | 3.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 4.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.4 | 2.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.4 | 3.5 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.4 | 6.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 3.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 2.7 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.3 | 11.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 9.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.3 | 1.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.3 | 7.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.3 | 7.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.5 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.3 | 4.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 4.7 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.3 | 12.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 0.8 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.3 | 3.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.3 | 1.1 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.3 | 4.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 1.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.3 | 1.8 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.2 | 6.0 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.2 | 0.7 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.2 | 10.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.7 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 0.7 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.2 | 8.9 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 3.3 | GO:0002638 | negative regulation of immunoglobulin production(GO:0002638) |
| 0.2 | 6.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 13.4 | GO:0032873 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.2 | 2.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 2.5 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.2 | 0.8 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 1.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 2.3 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.2 | 1.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.2 | 1.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.2 | 0.7 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 1.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.2 | 1.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.2 | 10.6 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.2 | 1.0 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 0.6 | GO:2000111 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.2 | 5.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 2.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 1.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.1 | 9.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 2.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.5 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 2.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 0.5 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 8.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.6 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 2.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 2.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 4.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 4.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 3.4 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 0.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.4 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 4.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 2.9 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.4 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 | 5.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 6.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.1 | 4.5 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.1 | 3.6 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 3.9 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 1.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.5 | GO:0006063 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) |
| 0.1 | 2.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.9 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.1 | 0.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.5 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.2 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 7.3 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 1.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 9.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.6 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 2.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.7 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 2.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.8 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 3.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 4.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.0 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 4.5 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 1.4 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.3 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 1.5 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 1.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 1.0 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 1.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 5.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 3.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 3.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.9 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 1.4 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 2.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.6 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.4 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 18.4 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.7 | 6.7 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 1.6 | 6.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 1.5 | 10.3 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 1.2 | 5.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 1.1 | 5.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.1 | 5.5 | GO:0071986 | Ragulator complex(GO:0071986) |
| 1.1 | 12.9 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 1.0 | 10.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.9 | 3.8 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.9 | 4.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.9 | 4.5 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.9 | 5.4 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.9 | 3.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.8 | 6.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.8 | 4.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.7 | 2.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 3.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.7 | 4.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.7 | 8.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.6 | 2.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.6 | 6.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.6 | 2.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.5 | 10.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.5 | 1.9 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.4 | 3.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 4.0 | GO:0034464 | BBSome(GO:0034464) |
| 0.4 | 3.4 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.4 | 8.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 4.7 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.4 | 6.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 7.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.4 | 2.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 1.9 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.3 | 2.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.3 | 1.0 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.3 | 8.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 1.8 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.3 | 5.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 2.3 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.3 | 5.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 5.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 2.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.3 | 5.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 4.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 6.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 3.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 7.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 4.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 1.3 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.2 | 3.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 11.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 4.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 1.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 18.1 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 1.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 1.7 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 4.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 10.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 8.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 5.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 3.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 7.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.3 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 5.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 5.5 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 11.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 5.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 6.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 5.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) filopodium tip(GO:0032433) |
| 0.1 | 2.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 3.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 4.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 9.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 4.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 3.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 10.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 17.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.0 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.9 | GO:0044439 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 10.8 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 14.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 4.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 3.6 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 2.1 | 8.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.4 | 5.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.3 | 3.8 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.2 | 10.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.2 | 9.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.2 | 12.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 1.1 | 6.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 1.0 | 10.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 1.0 | 10.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.9 | 7.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.8 | 4.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.8 | 3.8 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.7 | 3.7 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.7 | 2.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.7 | 6.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.7 | 3.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.7 | 3.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.7 | 3.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.7 | 3.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 7.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.7 | 2.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 4.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.6 | 1.8 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.6 | 17.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 5.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.6 | 4.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.5 | 2.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.5 | 3.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.5 | 1.5 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.5 | 1.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.5 | 10.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 8.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 6.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 16.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 1.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.4 | 1.6 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.4 | 5.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.4 | 2.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.4 | 5.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.4 | 2.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 16.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.4 | 10.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.4 | 3.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 1.0 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.3 | 3.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 0.9 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.2 | GO:0004793 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.3 | 1.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 4.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 1.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 3.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 0.8 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.3 | 3.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 3.8 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 1.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.7 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.2 | 2.3 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 6.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 7.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 1.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 6.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 14.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.2 | 2.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 1.7 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 3.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.6 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 2.9 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 4.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 4.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 1.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 9.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.2 | 2.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.2 | 1.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 2.6 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 2.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 3.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.6 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 1.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 6.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 2.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 2.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 7.0 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 3.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.9 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 3.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.5 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 4.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 4.7 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.1 | 4.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.4 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.1 | 11.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 2.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.4 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.0 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 1.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 1.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 19.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 3.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 1.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 4.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.1 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 1.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 1.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 3.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 4.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 12.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 19.2 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 11.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 3.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 13.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 9.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 3.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 1.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.3 | 20.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 13.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 27.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.2 | 10.0 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.2 | 6.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 11.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 3.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 19.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 4.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.9 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 7.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 12.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 3.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 5.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 6.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 4.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 5.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 22.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.7 | 6.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 5.9 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.6 | 9.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 10.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.5 | 1.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.5 | 4.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.5 | 8.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 5.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 7.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 9.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 10.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 2.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 4.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 3.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 16.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.3 | 23.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.3 | 7.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 1.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 8.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 3.7 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 12.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 6.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 6.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 8.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 6.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.2 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.2 | 10.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 4.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 9.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 6.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 5.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 2.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.5 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.9 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 3.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 3.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 10.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 5.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 6.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 5.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 2.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 5.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 6.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 2.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 1.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 2.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |