Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
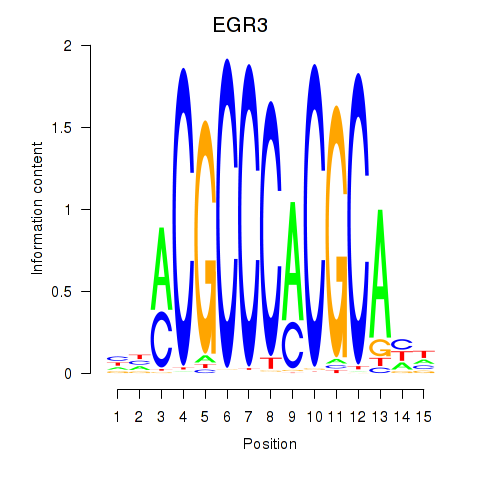
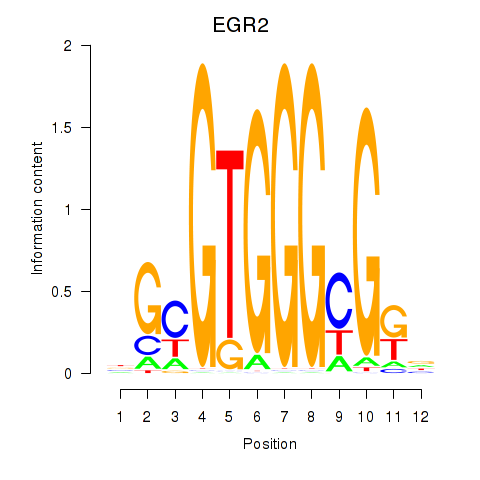
Results for EGR3_EGR2
Z-value: 0.15
Transcription factors associated with EGR3_EGR2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EGR3
|
ENSG00000179388.8 | early growth response 3 |
|
EGR2
|
ENSG00000122877.9 | early growth response 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EGR3 | hg19_v2_chr8_-_22550815_22550844 | 0.29 | 1.2e-05 | Click! |
| EGR2 | hg19_v2_chr10_-_64576105_64576133 | -0.19 | 4.7e-03 | Click! |
Activity profile of EGR3_EGR2 motif
Sorted Z-values of EGR3_EGR2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_124609823 | 36.04 |
ENST00000412681.2
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chr11_+_124609742 | 35.85 |
ENST00000284292.6
|
NRGN
|
neurogranin (protein kinase C substrate, RC3) |
| chrX_-_47479246 | 21.49 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr3_-_149688655 | 20.12 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr20_+_44035200 | 19.61 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr16_+_23847267 | 19.29 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr15_+_84115868 | 18.40 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr15_+_84116106 | 17.80 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr12_+_79258444 | 16.56 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr3_-_149688896 | 16.07 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr16_+_23847339 | 15.15 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr12_+_79258547 | 14.95 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr6_-_110500905 | 14.88 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr20_+_44034676 | 13.66 |
ENST00000372723.3
ENST00000372722.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr3_-_149688502 | 13.63 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chr19_+_35634146 | 13.54 |
ENST00000586063.1
ENST00000270310.2 ENST00000588265.1 |
FXYD7
|
FXYD domain containing ion transport regulator 7 |
| chr17_+_57697216 | 13.36 |
ENST00000393043.1
ENST00000269122.3 |
CLTC
|
clathrin, heavy chain (Hc) |
| chr14_+_100150622 | 12.94 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr4_+_83351715 | 12.24 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr4_+_158142750 | 11.91 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr4_+_83351791 | 11.79 |
ENST00000509635.1
|
ENOPH1
|
enolase-phosphatase 1 |
| chr17_-_42992856 | 11.77 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr4_-_90758118 | 11.73 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr15_-_71146480 | 11.60 |
ENST00000299213.8
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr11_-_125366089 | 11.42 |
ENST00000366139.3
ENST00000278919.3 |
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr5_+_67584174 | 11.13 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr6_-_110501200 | 10.95 |
ENST00000392586.1
ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1
|
WAS protein family, member 1 |
| chr17_+_44668035 | 10.48 |
ENST00000398238.4
ENST00000225282.8 |
NSF
|
N-ethylmaleimide-sensitive factor |
| chr14_-_23822080 | 10.35 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr19_-_36523709 | 10.28 |
ENST00000592017.1
ENST00000360535.4 |
CLIP3
|
CAP-GLY domain containing linker protein 3 |
| chr15_+_52311398 | 10.26 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr4_-_90758227 | 10.04 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr3_+_123813509 | 9.79 |
ENST00000460856.1
ENST00000240874.3 |
KALRN
|
kalirin, RhoGEF kinase |
| chr19_+_49617581 | 9.02 |
ENST00000391864.3
|
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr10_+_105036909 | 8.63 |
ENST00000369849.4
|
INA
|
internexin neuronal intermediate filament protein, alpha |
| chr4_-_153274078 | 8.31 |
ENST00000263981.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_+_30689401 | 8.26 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr1_+_6845384 | 8.26 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr14_+_100531615 | 8.22 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chrX_-_140271249 | 8.06 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr14_-_69445968 | 7.92 |
ENST00000438964.2
|
ACTN1
|
actinin, alpha 1 |
| chr12_-_96794330 | 7.86 |
ENST00000261211.3
|
CDK17
|
cyclin-dependent kinase 17 |
| chr12_-_49393092 | 7.61 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr6_+_30689350 | 7.40 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr7_+_95401851 | 7.37 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr19_-_13617037 | 7.36 |
ENST00000360228.5
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr8_-_101963677 | 7.23 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr19_+_49617609 | 7.07 |
ENST00000221459.2
ENST00000486217.2 |
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr7_+_95401877 | 7.01 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr8_-_101963482 | 6.97 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr10_-_101190202 | 6.97 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr14_-_102605983 | 6.95 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr10_-_75255724 | 6.95 |
ENST00000342558.3
ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr14_-_69445793 | 6.80 |
ENST00000538545.2
ENST00000394419.4 |
ACTN1
|
actinin, alpha 1 |
| chrX_+_47092314 | 6.77 |
ENST00000218348.3
|
USP11
|
ubiquitin specific peptidase 11 |
| chr4_-_83351294 | 6.72 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr14_-_69446034 | 6.56 |
ENST00000193403.6
|
ACTN1
|
actinin, alpha 1 |
| chr20_+_44034804 | 6.51 |
ENST00000357275.2
ENST00000372720.3 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr7_-_44365020 | 6.47 |
ENST00000395747.2
ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta |
| chr17_-_1083078 | 6.43 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr17_-_4890919 | 6.26 |
ENST00000572543.1
ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr3_+_35681081 | 6.24 |
ENST00000428373.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr2_+_187350883 | 6.20 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr2_+_187350973 | 6.20 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr3_+_238273 | 6.18 |
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr4_-_36246060 | 6.17 |
ENST00000303965.4
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr14_-_23770683 | 5.94 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr14_-_93651186 | 5.90 |
ENST00000556883.1
ENST00000298894.4 |
MOAP1
|
modulator of apoptosis 1 |
| chr15_+_91446157 | 5.83 |
ENST00000559717.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr9_+_131451480 | 5.81 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr6_-_84419101 | 5.71 |
ENST00000520302.1
ENST00000520213.1 ENST00000439399.2 ENST00000428679.2 ENST00000437520.1 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr19_+_10400615 | 5.61 |
ENST00000221980.4
|
ICAM5
|
intercellular adhesion molecule 5, telencephalin |
| chr11_+_63448955 | 5.59 |
ENST00000377819.5
ENST00000339997.4 ENST00000540798.1 ENST00000545432.1 ENST00000543552.1 ENST00000537981.1 |
RTN3
|
reticulon 3 |
| chr10_-_135150367 | 5.55 |
ENST00000368555.3
ENST00000252939.4 ENST00000368558.1 ENST00000368556.2 |
CALY
|
calcyon neuron-specific vesicular protein |
| chr11_+_63448918 | 5.53 |
ENST00000341307.2
ENST00000356000.3 ENST00000542238.1 |
RTN3
|
reticulon 3 |
| chr8_-_110703819 | 5.42 |
ENST00000532779.1
ENST00000534578.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr17_-_4890649 | 5.29 |
ENST00000361571.5
|
CAMTA2
|
calmodulin binding transcription activator 2 |
| chr12_+_110718921 | 5.28 |
ENST00000308664.6
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| chr13_+_42846272 | 5.23 |
ENST00000025301.2
|
AKAP11
|
A kinase (PRKA) anchor protein 11 |
| chr17_-_8066250 | 5.18 |
ENST00000488857.1
ENST00000481878.1 ENST00000316509.6 ENST00000498285.1 |
VAMP2
RP11-599B13.6
|
vesicle-associated membrane protein 2 (synaptobrevin 2) Uncharacterized protein |
| chr7_-_752577 | 5.16 |
ENST00000544935.1
ENST00000430040.1 ENST00000456696.2 ENST00000406797.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr2_+_191745535 | 5.16 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr8_-_110704014 | 5.14 |
ENST00000529190.1
ENST00000422135.1 ENST00000419099.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr8_-_101964265 | 4.96 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr9_+_33264861 | 4.95 |
ENST00000223500.8
|
CHMP5
|
charged multivesicular body protein 5 |
| chr1_-_32403903 | 4.95 |
ENST00000344035.6
ENST00000356536.3 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr15_+_80696666 | 4.95 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr5_-_132112907 | 4.94 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr5_-_132112921 | 4.91 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr16_+_5008290 | 4.90 |
ENST00000251170.7
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr11_-_122931881 | 4.83 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr9_+_130922537 | 4.74 |
ENST00000372994.1
|
C9orf16
|
chromosome 9 open reading frame 16 |
| chr3_-_171177852 | 4.67 |
ENST00000284483.8
ENST00000475336.1 ENST00000357327.5 ENST00000460047.1 ENST00000488470.1 ENST00000470834.1 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr19_-_51472031 | 4.60 |
ENST00000391808.1
|
KLK6
|
kallikrein-related peptidase 6 |
| chr2_+_191208196 | 4.50 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr9_+_19049372 | 4.48 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr5_-_88179302 | 4.45 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr9_-_15510989 | 4.38 |
ENST00000380715.1
ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr11_-_46867780 | 4.37 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr12_+_6875519 | 4.31 |
ENST00000389462.4
ENST00000540874.1 ENST00000309083.6 |
PTMS
|
parathymosin |
| chr2_-_27718052 | 4.26 |
ENST00000264703.3
|
FNDC4
|
fibronectin type III domain containing 4 |
| chr5_+_137774706 | 4.26 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr14_+_29234870 | 4.21 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr14_-_23822061 | 4.20 |
ENST00000397260.3
|
SLC22A17
|
solute carrier family 22, member 17 |
| chr4_-_102267953 | 4.19 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_+_86274145 | 4.17 |
ENST00000439827.1
ENST00000394720.2 ENST00000421579.1 |
GRM3
|
glutamate receptor, metabotropic 3 |
| chr10_+_95517660 | 4.10 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chrX_-_49056635 | 4.09 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr3_-_49907323 | 4.04 |
ENST00000296471.7
ENST00000488336.1 ENST00000467248.1 ENST00000466940.1 ENST00000463537.1 ENST00000480398.2 |
CAMKV
|
CaM kinase-like vesicle-associated |
| chr15_-_73925651 | 3.99 |
ENST00000545878.1
ENST00000287226.8 ENST00000345330.4 |
NPTN
|
neuroplastin |
| chr15_-_83316711 | 3.99 |
ENST00000568128.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_-_122932730 | 3.97 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr9_+_33265011 | 3.94 |
ENST00000419016.2
|
CHMP5
|
charged multivesicular body protein 5 |
| chr19_-_51472222 | 3.93 |
ENST00000376851.3
|
KLK6
|
kallikrein-related peptidase 6 |
| chr7_-_108096765 | 3.87 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr8_-_144242020 | 3.87 |
ENST00000414417.2
|
LY6H
|
lymphocyte antigen 6 complex, locus H |
| chr21_+_38445539 | 3.83 |
ENST00000418766.1
ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr3_-_52002403 | 3.81 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr19_+_40854559 | 3.79 |
ENST00000598962.1
ENST00000409419.1 ENST00000409587.1 ENST00000602131.1 ENST00000409735.4 ENST00000600948.1 ENST00000356508.5 ENST00000596682.1 ENST00000594908.1 |
PLD3
|
phospholipase D family, member 3 |
| chr6_-_112194484 | 3.79 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr4_+_41362796 | 3.75 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_+_102595119 | 3.68 |
ENST00000510890.1
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr12_-_65515334 | 3.68 |
ENST00000286574.4
|
WIF1
|
WNT inhibitory factor 1 |
| chr19_-_54693401 | 3.68 |
ENST00000338624.6
|
MBOAT7
|
membrane bound O-acyltransferase domain containing 7 |
| chr4_-_46391931 | 3.68 |
ENST00000381620.4
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr17_+_11924129 | 3.65 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr13_-_45010939 | 3.59 |
ENST00000261489.2
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr8_+_94929110 | 3.58 |
ENST00000520728.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr14_-_23772032 | 3.58 |
ENST00000452015.4
|
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr1_+_178995021 | 3.58 |
ENST00000263733.4
|
FAM20B
|
family with sequence similarity 20, member B |
| chr8_+_94929077 | 3.57 |
ENST00000297598.4
ENST00000520614.1 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr12_-_90102594 | 3.56 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr22_-_39268308 | 3.56 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr1_-_85156216 | 3.52 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr17_+_2496971 | 3.50 |
ENST00000397195.5
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
| chr9_-_139940608 | 3.50 |
ENST00000371601.4
|
NPDC1
|
neural proliferation, differentiation and control, 1 |
| chr7_-_108096822 | 3.49 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr22_-_38380543 | 3.49 |
ENST00000396884.2
|
SOX10
|
SRY (sex determining region Y)-box 10 |
| chr4_-_46391805 | 3.49 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr7_+_106685079 | 3.46 |
ENST00000265717.4
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta |
| chr7_+_153749732 | 3.45 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr7_+_24323782 | 3.42 |
ENST00000242152.2
ENST00000407573.1 |
NPY
|
neuropeptide Y |
| chr18_-_43684186 | 3.42 |
ENST00000590406.1
ENST00000282050.2 ENST00000590324.1 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr7_-_100493482 | 3.42 |
ENST00000411582.1
ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE
|
acetylcholinesterase (Yt blood group) |
| chr17_-_8534031 | 3.41 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr17_-_8534067 | 3.41 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr11_+_73882144 | 3.41 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr20_+_17207665 | 3.40 |
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr1_-_161087802 | 3.40 |
ENST00000368010.3
|
PFDN2
|
prefoldin subunit 2 |
| chr1_-_93426998 | 3.39 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr10_+_95517616 | 3.38 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr9_-_139922726 | 3.37 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr14_+_66975213 | 3.35 |
ENST00000543237.1
ENST00000305960.9 |
GPHN
|
gephyrin |
| chr10_+_95517566 | 3.35 |
ENST00000542308.1
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr1_-_9189229 | 3.33 |
ENST00000377411.4
|
GPR157
|
G protein-coupled receptor 157 |
| chr11_+_73882311 | 3.33 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr17_+_37026284 | 3.30 |
ENST00000433206.2
ENST00000435347.3 |
LASP1
|
LIM and SH3 protein 1 |
| chr12_+_49212514 | 3.29 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr17_-_8066843 | 3.29 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr1_-_153599732 | 3.24 |
ENST00000392623.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr5_-_142783694 | 3.23 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr19_+_45409011 | 3.23 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr3_-_32612263 | 3.23 |
ENST00000432458.2
ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr11_-_47447970 | 3.22 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr5_-_115177247 | 3.22 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr22_+_19701985 | 3.21 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr8_+_94929168 | 3.19 |
ENST00000518107.1
ENST00000396200.3 |
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_+_48796905 | 3.19 |
ENST00000505658.1
ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr19_-_13617247 | 3.18 |
ENST00000573710.2
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr9_+_33025209 | 3.17 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr1_-_153599426 | 3.17 |
ENST00000392622.1
|
S100A13
|
S100 calcium binding protein A13 |
| chr16_+_2564254 | 3.16 |
ENST00000565223.1
|
ATP6V0C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr1_-_154600421 | 3.15 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr10_-_118764862 | 3.15 |
ENST00000260777.10
|
KIAA1598
|
KIAA1598 |
| chr5_+_161274940 | 3.13 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr2_+_220306745 | 3.12 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr1_-_85156090 | 3.12 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr14_-_27066960 | 3.11 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr12_+_861717 | 3.05 |
ENST00000535572.1
|
WNK1
|
WNK lysine deficient protein kinase 1 |
| chr11_+_129939779 | 3.03 |
ENST00000533195.1
ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr12_+_118573663 | 3.01 |
ENST00000261313.2
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr1_+_110527308 | 2.98 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr16_+_2198604 | 2.97 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr1_-_23670817 | 2.95 |
ENST00000478691.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr6_+_44214824 | 2.93 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chrX_-_51812268 | 2.93 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr9_-_132597529 | 2.89 |
ENST00000372447.3
|
C9orf78
|
chromosome 9 open reading frame 78 |
| chr5_+_161274685 | 2.88 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr9_-_139922631 | 2.86 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr1_+_174846570 | 2.85 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr18_+_12948000 | 2.85 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr1_-_23670813 | 2.85 |
ENST00000374612.1
|
HNRNPR
|
heterogeneous nuclear ribonucleoprotein R |
| chr5_+_161275320 | 2.84 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_-_43833628 | 2.79 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr16_-_30134524 | 2.77 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr21_-_27542972 | 2.75 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr8_-_9760839 | 2.75 |
ENST00000519461.1
ENST00000517675.1 |
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr15_+_77223960 | 2.72 |
ENST00000394885.3
|
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr19_-_10530784 | 2.71 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr8_-_27468842 | 2.70 |
ENST00000523500.1
|
CLU
|
clusterin |
Network of associatons between targets according to the STRING database.
First level regulatory network of EGR3_EGR2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 34.4 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 10.5 | 31.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 4.8 | 58.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 4.4 | 21.8 | GO:1903285 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 3.2 | 25.8 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 3.2 | 3.2 | GO:0097114 | NMDA glutamate receptor clustering(GO:0097114) |
| 2.9 | 14.6 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.7 | 80.7 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 2.6 | 23.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 2.5 | 22.4 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 2.3 | 7.0 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 2.3 | 6.8 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 2.3 | 6.8 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 2.1 | 6.4 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 2.1 | 8.3 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 2.1 | 10.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 2.0 | 16.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 2.0 | 2.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 1.8 | 8.8 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 1.7 | 6.9 | GO:0043335 | protein unfolding(GO:0043335) |
| 1.7 | 8.3 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.6 | 4.9 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 1.6 | 9.8 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 1.6 | 4.8 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.5 | 13.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.4 | 21.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 1.4 | 4.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 1.4 | 4.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 1.3 | 4.0 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 1.3 | 21.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 1.3 | 14.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.3 | 11.7 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.2 | 6.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.2 | 2.5 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.2 | 7.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 1.2 | 3.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.2 | 8.5 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 1.2 | 3.6 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 1.1 | 3.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 1.1 | 5.6 | GO:0030070 | insulin processing(GO:0030070) |
| 1.1 | 3.2 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 1.0 | 7.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.9 | 5.6 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.9 | 6.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.9 | 0.9 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.9 | 9.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.9 | 8.1 | GO:0060137 | maternal process involved in parturition(GO:0060137) |
| 0.9 | 2.7 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.9 | 5.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.9 | 3.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.9 | 4.4 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.8 | 11.8 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.8 | 11.8 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.8 | 8.0 | GO:1900125 | regulation of hyaluronan biosynthetic process(GO:1900125) |
| 0.8 | 6.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.8 | 14.2 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.8 | 2.3 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.8 | 6.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.7 | 2.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.7 | 0.7 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.7 | 3.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 3.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.7 | 8.9 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.7 | 2.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.7 | 2.0 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.7 | 6.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 1.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.6 | 1.9 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.6 | 6.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.6 | 4.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.6 | 6.4 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.6 | 10.5 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.6 | 2.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.6 | 1.7 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.6 | 2.9 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.6 | 2.3 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.6 | 6.7 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.5 | 8.5 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.5 | 3.7 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.5 | 6.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.5 | 4.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.5 | 5.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.5 | 1.0 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.5 | 5.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 1.9 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.5 | 2.8 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 3.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.4 | 5.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 5.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.4 | 11.6 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.4 | 3.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 10.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 2.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 1.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.4 | 3.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.4 | 9.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.4 | 3.2 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.4 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.4 | 15.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 1.8 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 3.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.3 | 1.4 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.3 | 1.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 2.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.0 | GO:1900454 | positive regulation of norepinephrine secretion(GO:0010701) positive regulation of long term synaptic depression(GO:1900454) |
| 0.3 | 4.9 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.3 | 10.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 4.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 1.8 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.3 | 3.6 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.3 | 1.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.3 | 3.8 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.3 | 1.6 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.3 | 1.0 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.0 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.3 | 0.3 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.3 | 2.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.2 | 2.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.7 | GO:0090032 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) negative regulation of steroid hormone biosynthetic process(GO:0090032) |
| 0.2 | 1.2 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.2 | 0.7 | GO:1904796 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.2 | 2.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 1.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.2 | 1.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 0.5 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 0.7 | GO:0070541 | response to platinum ion(GO:0070541) cellular response to lead ion(GO:0071284) |
| 0.2 | 1.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 0.9 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.2 | 0.7 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.2 | 0.9 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.2 | 0.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.2 | 1.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 2.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.2 | 0.8 | GO:1901895 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.2 | 3.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 0.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 0.6 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.2 | 1.6 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 6.2 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 2.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.2 | 3.7 | GO:0043632 | modification-dependent macromolecule catabolic process(GO:0043632) |
| 0.2 | 8.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.2 | 0.8 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 0.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 13.1 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.2 | 4.7 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.2 | 0.6 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.2 | 3.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 1.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 2.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 12.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.2 | 3.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.2 | 0.9 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 0.7 | GO:0072183 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.2 | 1.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 1.0 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.2 | 2.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 2.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 3.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 2.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 0.5 | GO:0018201 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.2 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 3.5 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.2 | 3.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 2.5 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 0.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 2.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 6.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 3.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 4.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 1.9 | GO:0021756 | striatum development(GO:0021756) |
| 0.1 | 1.0 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 2.1 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.9 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.1 | 2.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 0.8 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 3.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.4 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 1.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 3.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.4 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.5 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 5.9 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.6 | GO:0021894 | cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.1 | 1.4 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.1 | 2.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.6 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 0.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.2 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.2 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.1 | 0.2 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 | 4.7 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.2 | GO:1902302 | regulation of heart rate by hormone(GO:0003064) regulation of potassium ion export(GO:1902302) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.4 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 2.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 0.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.1 | 1.6 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.1 | 0.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 13.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 2.7 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 1.9 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 1.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 2.6 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 1.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 6.3 | GO:0038096 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis(GO:0002433) Fc-gamma receptor signaling pathway involved in phagocytosis(GO:0038096) |
| 0.0 | 0.8 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 3.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.0 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 1.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 2.5 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.0 | 0.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 2.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.5 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 2.1 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.5 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.0 | 0.3 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 2.7 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.4 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 31.5 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 5.9 | 71.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 3.3 | 13.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 3.0 | 11.9 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 2.1 | 8.5 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.5 | 13.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 1.4 | 19.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 1.4 | 6.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.3 | 16.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 1.3 | 8.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.2 | 10.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 1.2 | 8.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.2 | 17.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.2 | 17.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.1 | 97.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 1.1 | 3.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 1.0 | 2.9 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.9 | 12.1 | GO:0097433 | dense body(GO:0097433) |
| 0.9 | 6.8 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.8 | 7.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 4.6 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.7 | 4.5 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.7 | 2.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.6 | 3.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.6 | 38.7 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.6 | 5.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.6 | 10.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 8.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.6 | 6.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.5 | 10.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 15.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 1.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 3.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.5 | 3.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 2.8 | GO:0035859 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.4 | 4.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 3.5 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.4 | 5.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 2.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.4 | 6.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.4 | 4.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 3.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.4 | 2.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 3.4 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.4 | 8.6 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.4 | 15.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 1.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 2.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 1.8 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.3 | 8.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 9.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 42.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.3 | 2.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 7.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 0.6 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.3 | 3.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 3.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 0.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.3 | 6.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.3 | 14.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.3 | 1.3 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.3 | 1.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 2.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 2.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 0.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 1.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 2.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 5.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 2.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.2 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.2 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 2.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 1.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 10.4 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 4.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 1.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 4.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 1.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 7.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 2.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 2.5 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.2 | 4.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 3.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 3.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 5.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 2.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 10.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 1.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 3.0 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 7.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 1.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 4.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 9.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 4.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 3.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 12.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 1.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 3.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 6.5 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 5.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 7.0 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 3.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 4.8 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 18.3 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 2.4 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.0 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.5 | 34.4 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 6.3 | 31.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.4 | 21.8 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 3.3 | 71.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 2.6 | 10.3 | GO:0097001 | ceramide binding(GO:0097001) |
| 2.3 | 7.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.0 | 18.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.7 | 10.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.5 | 16.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.5 | 11.7 | GO:0043559 | insulin binding(GO:0043559) |
| 1.3 | 16.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 1.2 | 4.9 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 1.2 | 6.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.2 | 4.8 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.2 | 30.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 1.1 | 12.2 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.1 | 10.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.0 | 8.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 1.0 | 46.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 1.0 | 4.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 1.0 | 11.9 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.0 | 9.9 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.0 | 2.9 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.9 | 2.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.9 | 7.0 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.9 | 3.5 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.9 | 4.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.8 | 3.3 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.8 | 4.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.8 | 15.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.8 | 16.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.8 | 3.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.8 | 2.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.8 | 4.6 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.7 | 16.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.7 | 9.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 1.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 12.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 3.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.6 | 7.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.6 | 2.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.6 | 7.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 12.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.5 | 3.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.5 | 3.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.5 | 3.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.5 | 1.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.5 | 10.2 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 7.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.5 | 3.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.5 | 6.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 3.9 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.5 | 2.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.4 | 4.5 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.4 | 4.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.4 | 9.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 1.2 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.4 | 10.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 11.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 5.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 8.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.4 | 4.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.3 | 4.5 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.3 | 1.4 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 11.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 1.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 4.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.3 | 21.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.3 | 3.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 4.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 4.9 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.3 | 2.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.3 | 3.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 3.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.3 | 5.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.3 | 0.9 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 3.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.3 | 1.1 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.3 | 2.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 1.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.3 | 5.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 12.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 11.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.3 | 2.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.2 | 15.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 3.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 4.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 6.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 0.6 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.2 | 5.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.9 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 2.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.2 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 0.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.5 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 2.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 0.9 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 4.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.8 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 0.7 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.2 | 2.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.5 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.2 | 2.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 3.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 5.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 1.1 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 21.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.8 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 1.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 2.9 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.2 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 2.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 6.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 3.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 21.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 4.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.5 | GO:0044388 | small protein activating enzyme binding(GO:0044388) RING-like zinc finger domain binding(GO:0071535) |
| 0.1 | 1.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 3.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 2.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 6.2 | GO:0015399 | primary active transmembrane transporter activity(GO:0015399) P-P-bond-hydrolysis-driven transmembrane transporter activity(GO:0015405) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.6 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 4.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.6 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 7.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.8 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 1.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 3.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 2.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 4.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 8.5 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 16.5 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 6.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 5.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 5.0 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 35.9 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 1.3 | 77.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.8 | 40.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.6 | 22.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.5 | 33.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 9.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.4 | 31.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.3 | 10.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 11.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 8.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 14.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 7.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 11.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 0.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 9.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 4.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 7.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 6.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 3.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 4.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 5.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 0.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 3.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 3.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 53.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 2.0 | 56.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.4 | 34.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 1.3 | 58.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 1.2 | 11.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.2 | 35.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 1.0 | 13.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.8 | 14.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.8 | 7.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.8 | 8.3 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.7 | 17.9 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.5 | 1.6 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.5 | 16.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 11.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.5 | 4.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 15.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 8.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.4 | 22.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 8.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.3 | 6.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 3.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 4.9 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 23.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.3 | 4.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.3 | 5.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 2.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 4.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 2.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 3.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.2 | 21.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 3.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 6.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 5.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 9.4 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.2 | 18.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 3.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 16.2 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 6.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 11.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.8 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.4 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 3.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 0.9 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.1 | 2.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.9 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 3.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 2.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 2.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 2.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 6.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |