Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
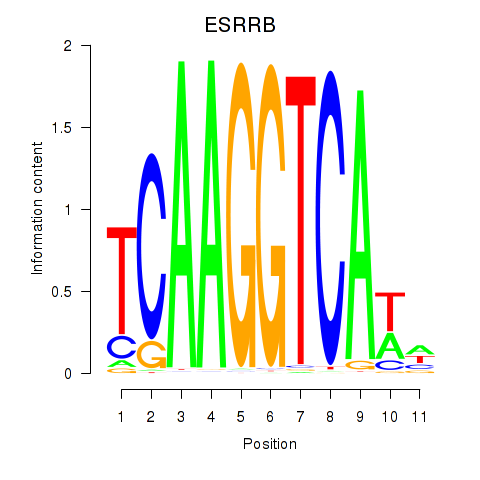
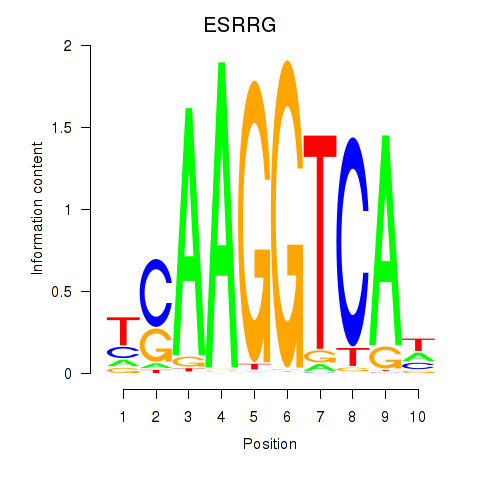
Results for ESRRB_ESRRG
Z-value: 0.68
Transcription factors associated with ESRRB_ESRRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESRRB
|
ENSG00000119715.10 | estrogen related receptor beta |
|
ESRRG
|
ENSG00000196482.12 | estrogen related receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESRRG | hg19_v2_chr1_-_217250231_217250349 | -0.40 | 5.3e-10 | Click! |
Activity profile of ESRRB_ESRRG motif
Sorted Z-values of ESRRB_ESRRG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_176046391 | 81.19 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr1_+_169077172 | 60.52 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr20_-_62130474 | 59.51 |
ENST00000217182.3
|
EEF1A2
|
eukaryotic translation elongation factor 1 alpha 2 |
| chr8_+_145149930 | 52.60 |
ENST00000318911.4
|
CYC1
|
cytochrome c-1 |
| chr14_-_103987679 | 48.19 |
ENST00000553610.1
|
CKB
|
creatine kinase, brain |
| chr10_+_81107271 | 45.45 |
ENST00000448165.1
|
PPIF
|
peptidylprolyl isomerase F |
| chr1_+_165600436 | 38.68 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr1_-_149889382 | 38.26 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr12_+_120875910 | 37.06 |
ENST00000551806.1
|
AL021546.6
|
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr7_-_30029367 | 34.60 |
ENST00000242059.5
|
SCRN1
|
secernin 1 |
| chr11_+_57480046 | 34.40 |
ENST00000378312.4
ENST00000278422.4 |
TMX2
|
thioredoxin-related transmembrane protein 2 |
| chr2_+_198365122 | 34.26 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr16_-_47177874 | 33.86 |
ENST00000562435.1
|
NETO2
|
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr10_+_81107216 | 33.21 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr14_-_21492251 | 33.16 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chr3_+_179322573 | 32.74 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr12_+_120875887 | 32.67 |
ENST00000229379.2
|
COX6A1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr17_+_79670386 | 32.58 |
ENST00000333676.3
ENST00000571730.1 ENST00000541223.1 |
MRPL12
SLC25A10
SLC25A10
|
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr7_+_128399002 | 32.47 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr15_+_43985725 | 31.81 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr12_+_6976687 | 31.67 |
ENST00000396705.5
|
TPI1
|
triosephosphate isomerase 1 |
| chr3_-_42845951 | 30.61 |
ENST00000418900.2
ENST00000430190.1 |
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr12_-_57039739 | 30.36 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr2_+_198365095 | 30.23 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr15_+_43985084 | 29.64 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr11_+_67798363 | 29.20 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_-_17380630 | 28.92 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr16_+_21964662 | 28.55 |
ENST00000561553.1
ENST00000565331.1 |
UQCRC2
|
ubiquinol-cytochrome c reductase core protein II |
| chr12_+_6643676 | 28.48 |
ENST00000396856.1
ENST00000396861.1 |
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr14_-_21492113 | 28.41 |
ENST00000554094.1
|
NDRG2
|
NDRG family member 2 |
| chr1_+_169075554 | 28.06 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_+_179322481 | 27.62 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr15_+_43885252 | 27.34 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr11_+_67798114 | 27.31 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr15_+_43886057 | 26.89 |
ENST00000441322.1
ENST00000413657.2 ENST00000453733.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr8_+_86376081 | 26.67 |
ENST00000285379.5
|
CA2
|
carbonic anhydrase II |
| chr9_-_130637244 | 26.52 |
ENST00000373156.1
|
AK1
|
adenylate kinase 1 |
| chr2_-_106054952 | 26.51 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr3_+_133465228 | 25.80 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr14_-_21491477 | 25.42 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr15_-_72523454 | 25.20 |
ENST00000565154.1
ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM
|
pyruvate kinase, muscle |
| chr10_-_101190202 | 25.17 |
ENST00000543866.1
ENST00000370508.5 |
GOT1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chrX_+_103031758 | 24.98 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr1_-_241683001 | 24.95 |
ENST00000366560.3
|
FH
|
fumarate hydratase |
| chr21_-_27107344 | 24.93 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr15_-_72523924 | 24.81 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr21_-_27107198 | 24.79 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr3_+_113465866 | 24.66 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr8_-_100905925 | 24.66 |
ENST00000518171.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr8_-_100905850 | 24.55 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr21_-_27107283 | 24.15 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr7_-_10979750 | 23.94 |
ENST00000339600.5
|
NDUFA4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr14_-_58893832 | 23.79 |
ENST00000556007.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr2_-_44223138 | 23.73 |
ENST00000260665.7
|
LRPPRC
|
leucine-rich pentatricopeptide repeat containing |
| chr17_-_29624343 | 23.63 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chrX_+_103031421 | 23.60 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr9_-_32573130 | 23.53 |
ENST00000350021.2
ENST00000379847.3 |
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr4_-_140216948 | 23.49 |
ENST00000265500.4
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr2_+_86426478 | 23.34 |
ENST00000254644.8
ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35
|
mitochondrial ribosomal protein L35 |
| chr11_+_34938119 | 23.34 |
ENST00000227868.4
ENST00000430469.2 ENST00000533262.1 |
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr22_-_39239987 | 23.01 |
ENST00000333039.2
|
NPTXR
|
neuronal pentraxin receptor |
| chr4_-_71705060 | 23.00 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr10_+_60028818 | 22.60 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr4_-_71705027 | 22.06 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr1_+_228270784 | 21.97 |
ENST00000541182.1
|
ARF1
|
ADP-ribosylation factor 1 |
| chr11_+_65770227 | 21.87 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr5_-_140027175 | 21.77 |
ENST00000512088.1
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr1_-_110284384 | 21.60 |
ENST00000540225.1
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr5_+_34757309 | 21.36 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr20_+_3776936 | 21.28 |
ENST00000439880.2
|
CDC25B
|
cell division cycle 25B |
| chr8_+_55047763 | 21.04 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr8_-_100905363 | 20.77 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chr2_+_98262497 | 20.62 |
ENST00000258424.2
|
COX5B
|
cytochrome c oxidase subunit Vb |
| chr3_-_10028366 | 20.59 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr18_-_43678241 | 20.35 |
ENST00000593152.2
ENST00000589252.1 ENST00000590665.1 ENST00000398752.6 |
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle |
| chr3_-_167452262 | 20.32 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chr12_+_6644443 | 19.96 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr11_-_790060 | 19.91 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr1_-_211752073 | 19.85 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr20_+_3776371 | 19.73 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr17_+_66509019 | 19.21 |
ENST00000585981.1
ENST00000589480.1 ENST00000585815.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr16_+_57481349 | 18.99 |
ENST00000262507.6
ENST00000565964.1 |
COQ9
|
coenzyme Q9 |
| chr2_-_220119280 | 18.91 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chrX_+_134166333 | 18.82 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr6_-_43027105 | 18.76 |
ENST00000230413.5
ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2
|
mitochondrial ribosomal protein L2 |
| chr5_+_218356 | 18.63 |
ENST00000264932.6
ENST00000504309.1 ENST00000510361.1 |
SDHA
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr1_-_26233423 | 18.26 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr16_+_56623433 | 18.11 |
ENST00000570176.1
|
MT3
|
metallothionein 3 |
| chr1_-_33502528 | 17.85 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chr16_-_58768177 | 17.80 |
ENST00000434819.2
ENST00000245206.5 |
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial |
| chr10_+_99185917 | 17.41 |
ENST00000334828.5
|
PGAM1
|
phosphoglycerate mutase 1 (brain) |
| chr2_+_220491973 | 17.25 |
ENST00000358055.3
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_+_19626531 | 17.02 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr15_+_78441663 | 16.92 |
ENST00000299518.2
ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A
|
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr17_-_27503770 | 16.91 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr1_-_205719295 | 16.85 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr11_+_65769946 | 16.41 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr2_-_86422095 | 16.31 |
ENST00000254636.5
|
IMMT
|
inner membrane protein, mitochondrial |
| chr16_+_58533951 | 16.16 |
ENST00000566192.1
ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4
|
NDRG family member 4 |
| chr16_-_47007545 | 15.88 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr16_-_19897455 | 15.87 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr16_+_4674787 | 15.85 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr7_+_44836276 | 15.67 |
ENST00000451562.1
ENST00000468812.1 ENST00000489459.1 ENST00000355968.6 |
PPIA
|
peptidylprolyl isomerase A (cyclophilin A) |
| chr1_-_153600656 | 15.60 |
ENST00000339556.4
ENST00000440685.2 |
S100A13
|
S100 calcium binding protein A13 |
| chr4_-_71705082 | 15.54 |
ENST00000439371.1
ENST00000499044.2 |
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chrX_-_13835147 | 15.49 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr3_-_98241760 | 15.49 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr8_+_124429006 | 15.21 |
ENST00000522194.1
ENST00000523356.1 |
WDYHV1
|
WDYHV motif containing 1 |
| chr1_+_153600869 | 15.05 |
ENST00000292169.1
ENST00000368696.3 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr19_-_12912601 | 14.95 |
ENST00000334482.5
|
PRDX2
|
peroxiredoxin 2 |
| chr22_+_29702996 | 14.86 |
ENST00000406549.3
ENST00000360113.2 ENST00000341313.6 ENST00000403764.1 ENST00000471961.1 ENST00000407854.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr4_+_41614909 | 14.62 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr16_-_81040415 | 14.58 |
ENST00000564174.1
ENST00000562713.1 ENST00000570195.1 ENST00000565925.1 ENST00000565108.1 ENST00000565650.1 ENST00000486645.1 |
CMC2
|
C-x(9)-C motif containing 2 |
| chr1_+_202995611 | 14.49 |
ENST00000367240.2
|
PPFIA4
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr20_+_30865429 | 14.46 |
ENST00000375712.3
|
KIF3B
|
kinesin family member 3B |
| chr12_-_53320245 | 14.28 |
ENST00000552150.1
|
KRT8
|
keratin 8 |
| chr15_-_66790146 | 14.22 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr7_+_56032270 | 14.18 |
ENST00000322090.3
ENST00000446778.1 |
GBAS
|
glioblastoma amplified sequence |
| chr5_-_133340326 | 14.03 |
ENST00000425992.1
ENST00000395044.3 ENST00000395047.2 |
VDAC1
|
voltage-dependent anion channel 1 |
| chr10_+_71078595 | 14.02 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr2_-_207024233 | 13.90 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr12_-_21810726 | 13.75 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr14_-_104387888 | 13.73 |
ENST00000286953.3
|
C14orf2
|
chromosome 14 open reading frame 2 |
| chrX_+_47053208 | 13.65 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr10_-_75634326 | 13.60 |
ENST00000322635.3
ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr8_-_131028869 | 13.52 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr8_-_110704014 | 13.44 |
ENST00000529190.1
ENST00000422135.1 ENST00000419099.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr19_-_29704448 | 13.36 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr19_+_17416609 | 13.35 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr8_+_110346546 | 13.34 |
ENST00000521662.1
ENST00000521688.1 ENST00000520147.1 |
ENY2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr12_-_21810765 | 13.29 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr11_+_118272328 | 13.00 |
ENST00000524422.1
|
ATP5L
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr3_-_98241358 | 12.94 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr12_+_119616447 | 12.93 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr13_-_48575401 | 12.84 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr16_+_4674814 | 12.73 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr20_+_34700333 | 12.41 |
ENST00000441639.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr19_+_35783028 | 12.37 |
ENST00000600291.1
ENST00000392213.3 |
MAG
|
myelin associated glycoprotein |
| chr14_-_104387831 | 12.31 |
ENST00000557040.1
ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2
|
chromosome 14 open reading frame 2 |
| chr5_-_137911049 | 12.24 |
ENST00000297185.3
|
HSPA9
|
heat shock 70kDa protein 9 (mortalin) |
| chr22_+_30163340 | 12.23 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr19_+_35783047 | 12.19 |
ENST00000595791.1
ENST00000597035.1 ENST00000537831.2 |
MAG
|
myelin associated glycoprotein |
| chr1_+_2036149 | 12.04 |
ENST00000482686.1
ENST00000400920.1 ENST00000486681.1 |
PRKCZ
|
protein kinase C, zeta |
| chr16_-_85833160 | 11.97 |
ENST00000435200.2
|
EMC8
|
ER membrane protein complex subunit 8 |
| chr16_-_85833109 | 11.93 |
ENST00000253457.3
|
EMC8
|
ER membrane protein complex subunit 8 |
| chr18_-_5544241 | 11.91 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr19_+_34855925 | 11.88 |
ENST00000590375.1
ENST00000356487.5 |
GPI
|
glucose-6-phosphate isomerase |
| chr19_+_35783037 | 11.86 |
ENST00000361922.4
|
MAG
|
myelin associated glycoprotein |
| chr19_+_34855874 | 11.81 |
ENST00000588991.2
|
GPI
|
glucose-6-phosphate isomerase |
| chrX_-_77225135 | 11.76 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr8_-_18541603 | 11.75 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr11_-_111957451 | 11.64 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr11_+_73498898 | 11.30 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr1_+_201924619 | 11.28 |
ENST00000367287.4
|
TIMM17A
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr3_-_167452298 | 11.17 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr19_+_34856141 | 11.06 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr6_-_34360413 | 10.95 |
ENST00000607016.1
|
NUDT3
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr16_-_3767551 | 10.86 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr16_-_3767506 | 10.50 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr6_-_97345689 | 10.46 |
ENST00000316149.7
|
NDUFAF4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr20_+_1875942 | 10.42 |
ENST00000358771.4
|
SIRPA
|
signal-regulatory protein alpha |
| chr16_-_19533404 | 10.38 |
ENST00000353258.3
|
GDE1
|
glycerophosphodiester phosphodiesterase 1 |
| chr21_-_35288284 | 10.38 |
ENST00000290299.2
|
ATP5O
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr12_+_98987369 | 10.29 |
ENST00000401722.3
ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
| chr1_-_241520385 | 10.27 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr1_-_241520525 | 10.26 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr7_-_123197733 | 10.20 |
ENST00000470123.1
ENST00000471770.1 |
NDUFA5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr20_+_57430162 | 10.16 |
ENST00000450130.1
ENST00000349036.3 ENST00000423897.1 |
GNAS
|
GNAS complex locus |
| chr1_+_84609944 | 10.15 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr16_-_15472151 | 9.96 |
ENST00000360151.4
ENST00000543801.1 |
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr7_+_48128194 | 9.96 |
ENST00000416681.1
ENST00000331803.4 ENST00000432131.1 |
UPP1
|
uridine phosphorylase 1 |
| chr9_+_131873227 | 9.88 |
ENST00000358994.4
ENST00000455292.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_-_43197189 | 9.73 |
ENST00000509253.1
ENST00000393987.2 ENST00000230431.6 |
DNPH1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr16_+_57481382 | 9.67 |
ENST00000564655.1
ENST00000567072.1 ENST00000567933.1 ENST00000563166.1 |
COQ9
|
coenzyme Q9 |
| chr15_-_41694640 | 9.64 |
ENST00000558719.1
ENST00000260361.4 ENST00000560978.1 |
NDUFAF1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr19_+_54926601 | 9.61 |
ENST00000301194.4
|
TTYH1
|
tweety family member 1 |
| chr7_+_48128816 | 9.60 |
ENST00000395564.4
|
UPP1
|
uridine phosphorylase 1 |
| chr3_+_52232102 | 9.58 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr19_+_54926621 | 9.53 |
ENST00000376530.3
ENST00000445095.1 ENST00000391739.3 ENST00000376531.3 |
TTYH1
|
tweety family member 1 |
| chr19_+_46850251 | 9.51 |
ENST00000012443.4
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr10_-_75634260 | 9.42 |
ENST00000372765.1
ENST00000351293.3 |
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr7_+_48128316 | 9.41 |
ENST00000341253.4
|
UPP1
|
uridine phosphorylase 1 |
| chr2_+_220492116 | 9.34 |
ENST00000373760.2
|
SLC4A3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_+_46850320 | 9.33 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr7_-_43965937 | 9.27 |
ENST00000455877.1
ENST00000223341.7 ENST00000447717.3 ENST00000426198.1 |
URGCP
|
upregulator of cell proliferation |
| chr3_+_184038073 | 9.26 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr16_+_6069586 | 9.17 |
ENST00000547372.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr2_-_207023918 | 9.12 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr4_-_74088800 | 9.11 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr2_+_192141611 | 9.10 |
ENST00000392316.1
|
MYO1B
|
myosin IB |
| chr17_-_42144949 | 9.07 |
ENST00000591247.1
|
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr7_+_48128854 | 9.06 |
ENST00000436673.1
ENST00000429491.2 |
UPP1
|
uridine phosphorylase 1 |
| chr7_+_153584166 | 9.01 |
ENST00000404039.1
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr17_-_40729681 | 8.97 |
ENST00000590760.1
ENST00000587209.1 ENST00000393795.3 ENST00000253789.5 |
PSMC3IP
|
PSMC3 interacting protein |
| chr14_-_21490958 | 8.88 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr3_-_183735731 | 8.78 |
ENST00000334444.6
|
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_-_207024134 | 8.70 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr22_-_43036607 | 8.62 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr20_-_3748416 | 8.57 |
ENST00000399672.1
|
C20orf27
|
chromosome 20 open reading frame 27 |
| chr11_-_57298187 | 8.50 |
ENST00000525158.1
ENST00000257245.4 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESRRB_ESRRG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.2 | 78.7 | GO:2000276 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 22.6 | 67.9 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 22.1 | 88.6 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 16.1 | 48.4 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 13.4 | 40.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 12.0 | 48.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 11.6 | 34.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 10.6 | 31.7 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 10.1 | 322.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 9.5 | 38.0 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 8.9 | 26.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 8.3 | 50.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 8.2 | 24.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 7.6 | 22.7 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 7.5 | 22.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 7.3 | 29.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 7.2 | 7.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 7.2 | 143.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 7.1 | 28.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 6.4 | 63.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 6.3 | 31.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 6.0 | 95.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 5.9 | 23.7 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 5.6 | 16.9 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 5.6 | 11.2 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 5.3 | 21.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 5.3 | 26.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 4.9 | 29.1 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 4.7 | 47.5 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 4.6 | 18.3 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 4.4 | 301.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 4.3 | 25.8 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 4.3 | 38.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 4.3 | 59.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 4.2 | 16.8 | GO:0019046 | release from viral latency(GO:0019046) |
| 4.1 | 41.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 4.0 | 19.9 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 4.0 | 19.8 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 3.7 | 18.5 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 3.7 | 85.0 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 3.2 | 12.9 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 3.2 | 38.3 | GO:0015074 | DNA integration(GO:0015074) |
| 3.1 | 18.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 3.1 | 15.5 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 3.1 | 21.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 3.1 | 12.2 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 2.9 | 23.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 2.9 | 11.6 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 2.8 | 19.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 2.7 | 29.9 | GO:0042407 | cristae formation(GO:0042407) |
| 2.6 | 25.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 2.4 | 28.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 2.4 | 7.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 2.3 | 13.8 | GO:1903027 | regulation of opsonization(GO:1903027) |
| 2.3 | 9.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 2.2 | 10.9 | GO:1901908 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 2.0 | 10.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 2.0 | 12.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 2.0 | 14.0 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 2.0 | 14.0 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 2.0 | 11.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.9 | 13.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 1.9 | 5.6 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.9 | 14.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.9 | 5.6 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.8 | 12.9 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.8 | 9.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.8 | 9.0 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 1.8 | 5.3 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.6 | 22.6 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 1.6 | 49.4 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 1.6 | 15.6 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 1.5 | 6.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.5 | 12.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 1.5 | 30.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.5 | 4.5 | GO:1902722 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) positive regulation of prolactin secretion(GO:1902722) |
| 1.5 | 3.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 1.5 | 7.3 | GO:0060356 | leucine import(GO:0060356) |
| 1.5 | 23.3 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 1.5 | 33.4 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 1.5 | 10.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 1.4 | 5.7 | GO:0030242 | pexophagy(GO:0030242) |
| 1.4 | 5.6 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 1.4 | 101.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 1.4 | 19.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 1.3 | 13.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 1.3 | 13.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 1.3 | 24.6 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 1.3 | 20.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 1.3 | 16.6 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.3 | 15.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 1.3 | 13.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 1.2 | 14.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 1.2 | 18.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 1.2 | 5.9 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 1.2 | 24.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 1.2 | 14.0 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 1.2 | 16.2 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 1.1 | 5.5 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 1.1 | 3.3 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.0 | 14.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 1.0 | 6.9 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 1.0 | 4.9 | GO:1903275 | regulation of sodium ion export(GO:1903273) positive regulation of sodium ion export(GO:1903275) regulation of sodium ion export from cell(GO:1903276) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.9 | 55.0 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.9 | 5.6 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.9 | 5.4 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.9 | 9.7 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.9 | 7.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.9 | 5.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.8 | 2.4 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.8 | 5.5 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.8 | 43.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.8 | 7.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.8 | 6.9 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.8 | 7.5 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.7 | 9.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.7 | 4.1 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.7 | 3.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.6 | 8.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.6 | 14.5 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.6 | 8.8 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.6 | 29.5 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.6 | 3.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.6 | 28.7 | GO:1900449 | regulation of neurotransmitter receptor activity(GO:0099601) regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.6 | 14.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.6 | 15.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.6 | 2.8 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.5 | 10.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.5 | 3.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.5 | 2.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 12.9 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.4 | 8.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 11.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.4 | 0.9 | GO:0052552 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.4 | 5.7 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.4 | 5.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.4 | 1.6 | GO:0045915 | response to selenium ion(GO:0010269) positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.4 | 14.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.4 | 1.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.4 | 4.8 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.4 | 2.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 | 1.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.4 | 7.1 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 3.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.3 | 1.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 42.4 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.3 | 24.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.3 | 2.6 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 20.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.3 | 3.0 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.3 | 2.7 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.3 | 2.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 9.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.3 | 5.5 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.3 | 5.8 | GO:0002192 | IRES-dependent translational initiation(GO:0002192) |
| 0.3 | 6.6 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.3 | 0.8 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) positive regulation of telomeric DNA binding(GO:1904744) |
| 0.3 | 2.4 | GO:0007613 | memory(GO:0007613) |
| 0.3 | 8.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 14.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.3 | 53.7 | GO:0006457 | protein folding(GO:0006457) |
| 0.2 | 7.4 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.2 | 2.9 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.2 | 2.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 3.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 5.2 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.2 | 3.8 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 1.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 6.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 17.8 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.2 | 3.9 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 2.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 1.8 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.2 | 2.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 1.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) protein localization to nucleolus(GO:1902570) |
| 0.2 | 6.0 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 1.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.2 | 1.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 6.6 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.2 | 3.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.2 | 7.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 2.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 7.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 1.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 3.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 2.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 3.6 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 9.0 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 4.1 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 2.7 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.1 | 6.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 8.9 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 2.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 2.2 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 14.6 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.9 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.9 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 0.5 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 2.8 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.9 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.7 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 1.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.7 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 4.3 | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation(GO:0050731) |
| 0.0 | 2.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.0 | 1.5 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 1.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 4.7 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.0 | 0.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.7 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.9 | 51.4 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 12.1 | 36.4 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 11.0 | 197.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 10.3 | 144.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 10.0 | 139.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 7.2 | 64.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 6.9 | 48.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 6.3 | 31.7 | GO:0070470 | plasma membrane respiratory chain complex I(GO:0045272) plasma membrane respiratory chain(GO:0070470) |
| 6.0 | 59.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 5.6 | 5.6 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 5.5 | 43.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 4.9 | 44.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 4.8 | 14.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 4.7 | 93.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 4.3 | 25.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 4.0 | 240.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 3.3 | 13.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 3.2 | 128.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 3.0 | 21.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 3.0 | 41.5 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 3.0 | 32.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.7 | 18.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 2.6 | 23.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 2.3 | 9.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 2.3 | 38.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 2.1 | 109.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 2.0 | 21.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 1.9 | 19.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 1.9 | 24.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.9 | 33.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 1.9 | 5.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 1.8 | 18.1 | GO:0097449 | astrocyte projection(GO:0097449) |
| 1.6 | 18.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 1.6 | 29.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.5 | 350.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.5 | 4.5 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 1.5 | 13.4 | GO:0097433 | dense body(GO:0097433) |
| 1.4 | 7.2 | GO:0032021 | NELF complex(GO:0032021) |
| 1.4 | 5.7 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 1.4 | 16.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.3 | 12.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 1.3 | 11.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.3 | 25.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 1.3 | 5.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.2 | 8.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.1 | 24.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 1.1 | 2.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 1.0 | 3.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 1.0 | 7.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 1.0 | 5.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.9 | 13.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.9 | 2.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.9 | 3.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.9 | 11.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.9 | 36.6 | GO:0031430 | M band(GO:0031430) |
| 0.8 | 6.8 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.8 | 6.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.8 | 49.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.7 | 37.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.7 | 15.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 5.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.6 | 3.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.5 | 143.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.5 | 13.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.5 | 13.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.5 | 15.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.4 | 16.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 79.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.4 | 11.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 3.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.4 | 2.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 3.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 11.8 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.3 | 25.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 4.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.3 | 4.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 2.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 10.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 32.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 3.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 21.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.2 | 65.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.2 | 29.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 0.5 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 21.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 12.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 38.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 20.4 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.1 | 6.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 7.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 17.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 1.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 29.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 4.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 5.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 5.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.1 | 9.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 5.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 13.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 4.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.2 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.5 | 52.6 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 17.1 | 51.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 14.3 | 43.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 14.0 | 42.0 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 14.0 | 111.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 12.5 | 50.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 12.1 | 48.4 | GO:0019828 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 11.6 | 34.7 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 9.5 | 38.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 8.5 | 50.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 7.6 | 22.7 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 7.3 | 22.0 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 7.3 | 29.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 7.0 | 97.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 6.0 | 138.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 5.8 | 167.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 5.5 | 88.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 5.1 | 262.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 4.3 | 12.8 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 4.3 | 51.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 4.2 | 16.9 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 4.0 | 48.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 3.9 | 27.0 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 3.7 | 25.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 3.3 | 26.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 3.3 | 16.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 3.3 | 19.8 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 3.2 | 9.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 3.2 | 19.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 3.1 | 24.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 2.9 | 23.3 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 2.6 | 36.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 2.6 | 10.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 2.6 | 10.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 2.5 | 24.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 2.4 | 43.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 2.3 | 13.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 2.3 | 61.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 2.3 | 18.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 2.2 | 10.9 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 2.1 | 29.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.9 | 14.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 1.9 | 20.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.8 | 5.3 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 1.8 | 14.0 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.7 | 15.0 | GO:0015288 | porin activity(GO:0015288) |
| 1.5 | 3.0 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 1.5 | 70.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 1.5 | 87.6 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 1.4 | 11.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 1.4 | 8.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.4 | 5.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.3 | 20.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 1.3 | 5.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 1.2 | 18.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 1.2 | 32.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 1.2 | 4.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.2 | 6.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 1.2 | 7.0 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.2 | 3.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.1 | 8.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 1.0 | 4.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 1.0 | 7.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.0 | 19.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 1.0 | 19.7 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.0 | 5.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.9 | 10.2 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.9 | 5.5 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.9 | 4.5 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 96.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.9 | 2.7 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.9 | 2.6 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.9 | 26.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.8 | 5.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.8 | 18.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.8 | 10.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.8 | 15.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.8 | 15.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.7 | 8.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.7 | 7.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.7 | 25.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.6 | 8.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.6 | 3.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.6 | 9.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 14.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.5 | 10.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.5 | 3.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.5 | 5.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.5 | 12.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.5 | 2.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 2.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.5 | 8.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.5 | 4.8 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.5 | 4.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 54.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.5 | 2.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.5 | 14.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 6.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.4 | 1.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.4 | 24.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.4 | 2.9 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.4 | 9.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 25.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.4 | 83.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.4 | 5.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 3.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.4 | 2.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.3 | 4.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.3 | 9.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.3 | 3.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 7.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 41.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.3 | 7.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.3 | 3.0 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 1.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 63.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 3.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.9 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.2 | 28.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 1.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 6.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.2 | 2.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 8.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 7.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 14.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 3.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 17.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 6.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 7.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 2.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 24.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 0.8 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 1.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 8.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 3.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 6.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 6.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.3 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 5.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 3.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 41.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 1.3 | 106.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 1.1 | 41.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.0 | 18.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.9 | 22.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.9 | 11.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.9 | 26.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.8 | 101.0 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.8 | 113.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.7 | 16.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.7 | 81.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 7.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 19.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 3.0 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 3.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 9.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 10.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 11.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 3.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.2 | 3.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.2 | 3.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 6.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 7.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 26.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 5.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 168.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 6.7 | 570.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 6.0 | 54.0 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 3.2 | 132.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 3.2 | 208.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 3.0 | 54.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 2.3 | 53.5 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 2.2 | 22.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 2.0 | 63.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.8 | 26.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 1.8 | 83.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.6 | 54.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.5 | 41.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 1.5 | 20.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 1.4 | 12.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.3 | 45.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 1.3 | 26.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 1.2 | 19.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 1.0 | 18.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.9 | 13.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.8 | 7.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.8 | 19.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.7 | 18.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.6 | 7.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 14.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.6 | 155.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.5 | 4.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.4 | 16.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.4 | 10.2 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.4 | 9.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.4 | 5.4 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.4 | 9.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 5.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 6.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 31.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 8.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 56.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.3 | 27.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.3 | 7.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.3 | 5.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 4.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 11.2 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 2.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 3.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 10.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 3.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 8.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 3.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.2 | 4.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 9.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 4.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 5.3 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 16.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 3.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 2.0 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 15.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.9 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 1.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 3.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 5.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 2.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.8 | REACTOME MITOTIC M M G1 PHASES | Genes involved in Mitotic M-M/G1 phases |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |