Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
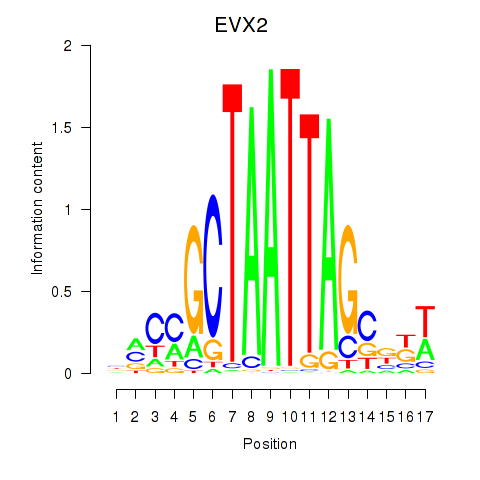
Results for EVX2
Z-value: 0.07
Transcription factors associated with EVX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EVX2
|
ENSG00000174279.4 | even-skipped homeobox 2 |
Activity profile of EVX2 motif
Sorted Z-values of EVX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_111718173 | 13.73 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717600 | 13.63 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111718036 | 13.47 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717511 | 13.17 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr6_+_39760129 | 12.18 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr3_+_115342349 | 9.09 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr11_-_115375107 | 7.70 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr17_-_9929581 | 7.41 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr17_-_27418537 | 2.77 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr1_-_151431909 | 2.73 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr1_-_151431647 | 2.29 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr18_-_33709268 | 2.03 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr12_+_15699286 | 1.46 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr4_-_41884620 | 1.01 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr6_-_62996066 | 0.99 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr12_+_28410128 | 0.76 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_+_28261533 | 0.71 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr19_+_11071546 | 0.43 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr6_-_111927062 | 0.40 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr6_-_111927449 | 0.27 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr17_-_40337470 | 0.16 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr15_-_55563072 | 0.13 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_+_28261492 | 0.11 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr1_+_28261621 | 0.10 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr15_-_55562479 | 0.07 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr15_-_55562582 | 0.02 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.9 | 9.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.5 | 1.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.4 | 5.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 2.0 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 12.2 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0007068 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.8 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 52.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.7 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 7.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 9.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 54.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 7.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 5.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 54.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 12.2 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 2.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 7.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |