Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
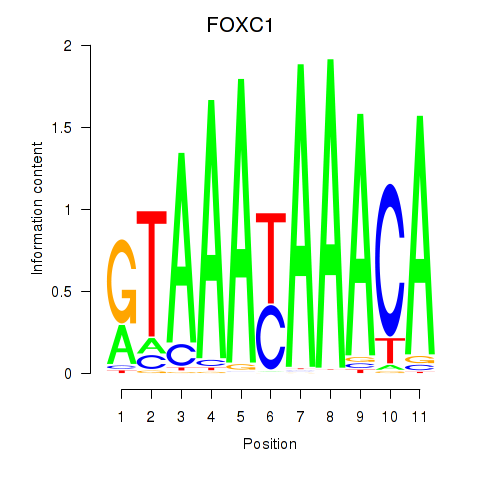
Results for FOXC1
Z-value: 0.67
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | -0.12 | 8.6e-02 | Click! |
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_34204642 | 11.93 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr10_+_114709999 | 11.87 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_76462713 | 9.81 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr10_+_114710211 | 9.51 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_152146385 | 8.82 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr5_-_111091948 | 8.82 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr4_-_102267953 | 8.31 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr7_-_99698338 | 8.21 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr10_+_54074033 | 7.59 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr3_-_49066811 | 7.44 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr12_-_49333446 | 7.24 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr1_-_150669500 | 7.24 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr2_+_181845843 | 7.23 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr4_-_102268628 | 7.20 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_+_16109519 | 7.17 |
ENST00000526530.1
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr8_-_80993010 | 6.84 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr13_+_52598827 | 6.80 |
ENST00000521776.2
|
UTP14C
|
UTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) |
| chr4_-_102268484 | 6.62 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_+_10748019 | 6.54 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr8_+_11666649 | 6.51 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_225434538 | 6.22 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr6_+_10747986 | 6.11 |
ENST00000379542.5
|
TMEM14B
|
transmembrane protein 14B |
| chr8_-_23261589 | 5.89 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr6_-_42016385 | 5.80 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr1_-_19811132 | 5.75 |
ENST00000433834.1
|
CAPZB
|
capping protein (actin filament) muscle Z-line, beta |
| chr22_-_43036607 | 5.62 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr6_+_119215308 | 5.58 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr12_-_123756781 | 5.37 |
ENST00000544658.1
|
CDK2AP1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr12_-_31479045 | 5.30 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr6_-_79787902 | 4.89 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr4_-_105416039 | 4.85 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr12_-_92539614 | 4.80 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr4_+_147096837 | 4.74 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_151431647 | 4.63 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr2_-_207023918 | 4.56 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr7_-_27219849 | 4.46 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr5_+_159436120 | 4.38 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr4_-_83765613 | 4.28 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr1_-_94079648 | 4.25 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr8_+_54764346 | 4.15 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr3_+_171758344 | 4.01 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr15_+_96869165 | 4.00 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr7_-_95064264 | 4.00 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr2_-_207024134 | 3.94 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr11_-_108464465 | 3.91 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr2_-_190044480 | 3.90 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr12_+_27175476 | 3.87 |
ENST00000546323.1
ENST00000282892.3 |
MED21
|
mediator complex subunit 21 |
| chr17_-_46688334 | 3.87 |
ENST00000239165.7
|
HOXB7
|
homeobox B7 |
| chr20_-_62582475 | 3.82 |
ENST00000369908.5
|
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr8_-_141931287 | 3.81 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr16_+_30064444 | 3.80 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_30064411 | 3.75 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_58655461 | 3.71 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr10_+_13652047 | 3.66 |
ENST00000601460.1
|
RP11-295P9.3
|
Uncharacterized protein |
| chr1_+_226411319 | 3.56 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr6_-_5260963 | 3.49 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chr7_-_86849025 | 3.47 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr3_+_142315225 | 3.34 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr5_-_39425068 | 3.32 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr20_-_62587735 | 3.23 |
ENST00000354216.6
ENST00000369892.3 ENST00000358711.3 |
UCKL1
|
uridine-cytidine kinase 1-like 1 |
| chr3_-_168865522 | 3.09 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_-_43855444 | 3.06 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr5_-_39425222 | 3.02 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_+_12524965 | 2.96 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr22_-_39268308 | 2.84 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr4_-_164534657 | 2.78 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr11_-_66112555 | 2.73 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr5_-_39425290 | 2.73 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr10_+_71561649 | 2.73 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr15_+_80351910 | 2.72 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr8_-_93107443 | 2.65 |
ENST00000360348.2
ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_151431909 | 2.65 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr15_+_80351977 | 2.62 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr17_+_79953310 | 2.56 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr4_-_103266626 | 2.54 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr5_+_179921430 | 2.53 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr1_+_73771844 | 2.52 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr5_+_112312416 | 2.51 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr2_-_72375167 | 2.46 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr3_-_11623804 | 2.45 |
ENST00000451674.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr10_-_118032979 | 2.40 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr3_-_128902759 | 2.39 |
ENST00000422453.2
ENST00000504813.1 ENST00000512338.1 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr10_-_118032697 | 2.35 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr8_+_31497271 | 2.30 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr2_-_207024233 | 2.18 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_25664408 | 2.18 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr7_-_14028488 | 2.16 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr5_+_179921344 | 2.13 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr11_-_108464321 | 2.10 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr3_-_141868293 | 2.04 |
ENST00000317104.7
ENST00000494358.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_-_111291587 | 2.00 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr18_+_21032781 | 1.94 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr3_-_16524357 | 1.93 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr17_+_57642886 | 1.89 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr12_+_54378923 | 1.85 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr2_+_28974668 | 1.84 |
ENST00000296122.6
ENST00000395366.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_-_99578776 | 1.81 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr1_-_236767779 | 1.80 |
ENST00000366579.1
ENST00000366582.3 ENST00000366581.2 |
HEATR1
|
HEAT repeat containing 1 |
| chr10_+_71561704 | 1.79 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr6_+_134274322 | 1.77 |
ENST00000367871.1
ENST00000237264.4 |
TBPL1
|
TBP-like 1 |
| chr11_+_93479588 | 1.73 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr2_+_33359646 | 1.73 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr10_-_118031778 | 1.72 |
ENST00000369236.1
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr2_+_33359687 | 1.59 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr9_-_136933134 | 1.58 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr3_+_69812877 | 1.54 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr7_-_14029283 | 1.52 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr3_-_128902729 | 1.48 |
ENST00000451728.2
ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP
|
CCHC-type zinc finger, nucleic acid binding protein |
| chr15_+_67418047 | 1.46 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr17_+_41363854 | 1.43 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr8_+_97597148 | 1.41 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr1_-_246729544 | 1.40 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr2_-_27886460 | 1.40 |
ENST00000404798.2
ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr8_-_93107696 | 1.39 |
ENST00000436581.2
ENST00000520583.1 ENST00000519061.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr4_-_89744457 | 1.38 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr15_+_49462434 | 1.37 |
ENST00000558145.1
ENST00000543495.1 ENST00000544523.1 ENST00000560138.1 |
GALK2
|
galactokinase 2 |
| chr10_+_71561630 | 1.36 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_+_27886330 | 1.35 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr16_+_56969284 | 1.31 |
ENST00000568358.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr7_+_120590803 | 1.31 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr3_+_48481658 | 1.30 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog (S. cerevisiae) |
| chr11_-_104480019 | 1.26 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr6_+_10528560 | 1.25 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr10_+_95517566 | 1.25 |
ENST00000542308.1
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr14_+_23340822 | 1.23 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr6_-_31651817 | 1.21 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr15_-_30114622 | 1.18 |
ENST00000495972.2
ENST00000346128.6 |
TJP1
|
tight junction protein 1 |
| chr2_+_28974603 | 1.13 |
ENST00000441461.1
ENST00000358506.2 |
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_94312706 | 1.02 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr7_-_72972319 | 0.99 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr6_-_52926539 | 0.95 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr3_-_48481518 | 0.88 |
ENST00000412398.2
ENST00000395696.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr1_-_159684371 | 0.87 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr13_+_98086445 | 0.82 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr3_-_24207039 | 0.81 |
ENST00000280696.5
|
THRB
|
thyroid hormone receptor, beta |
| chr3_-_114477962 | 0.78 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chrX_+_9431324 | 0.78 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr3_-_114477787 | 0.78 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr19_-_4831701 | 0.77 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr4_-_185395672 | 0.73 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr10_+_105314881 | 0.70 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr6_+_30130969 | 0.66 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr10_+_95517616 | 0.64 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr17_-_37934466 | 0.61 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 (Aiolos) |
| chr18_-_53070913 | 0.60 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr3_-_127455200 | 0.58 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr8_-_95449155 | 0.57 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr12_+_59989918 | 0.56 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr18_+_19749386 | 0.55 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr3_-_48481434 | 0.55 |
ENST00000395694.2
ENST00000447018.1 ENST00000442740.1 |
CCDC51
|
coiled-coil domain containing 51 |
| chr1_+_32739733 | 0.54 |
ENST00000333070.4
|
LCK
|
lymphocyte-specific protein tyrosine kinase |
| chr3_-_141868357 | 0.52 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_-_74486347 | 0.38 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_+_134103496 | 0.36 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr18_-_53303123 | 0.32 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr4_-_99578789 | 0.30 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr17_+_72426891 | 0.26 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr8_-_93107827 | 0.22 |
ENST00000520724.1
ENST00000518844.1 |
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_-_133969673 | 0.21 |
ENST00000427044.2
|
RYK
|
receptor-like tyrosine kinase |
| chr12_-_68696652 | 0.16 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr10_+_95517660 | 0.15 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr2_+_207024306 | 0.13 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr7_+_136553370 | 0.11 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr1_-_109935819 | 0.09 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr8_+_70404996 | 0.09 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr8_+_120079478 | 0.05 |
ENST00000332843.2
|
COLEC10
|
collectin sub-family member 10 (C-type lectin) |
| chr5_+_150040403 | 0.05 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr20_-_22566089 | 0.04 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr7_+_80231466 | 0.04 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_+_105315102 | 0.03 |
ENST00000369777.2
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr3_+_40351169 | 0.02 |
ENST00000232905.3
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr3_-_57233966 | 0.02 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 22.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 3.0 | 11.9 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 2.9 | 8.8 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 2.9 | 11.5 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 2.4 | 14.3 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 2.3 | 9.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.8 | 3.6 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 1.6 | 6.2 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 1.3 | 6.5 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 1.2 | 7.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.2 | 7.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 1.2 | 7.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 1.1 | 5.7 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 1.0 | 4.0 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.9 | 8.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.8 | 2.5 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.8 | 4.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.8 | 4.7 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.7 | 3.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.6 | 5.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.6 | 1.8 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.6 | 6.6 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.6 | 7.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 0.5 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.4 | 1.8 | GO:0006235 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.4 | 3.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 7.6 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 1.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 2.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.3 | 1.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 6.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 3.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.3 | 1.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 1.9 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.3 | 0.8 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.3 | 2.3 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 2.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 2.7 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.0 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 2.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 0.7 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 4.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 4.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 3.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 5.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 1.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 1.4 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 10.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.2 | 1.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 1.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 4.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.2 | 5.9 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 4.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.2 | 7.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 10.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 3.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 3.9 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.8 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 5.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 3.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 5.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 0.8 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.1 | 4.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.6 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.9 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.1 | 9.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 5.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 1.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 3.9 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.1 | 2.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 3.9 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.5 | GO:0050862 | cellular zinc ion homeostasis(GO:0006882) positive regulation of T cell receptor signaling pathway(GO:0050862) zinc ion homeostasis(GO:0055069) |
| 0.0 | 1.5 | GO:2000144 | melanocyte differentiation(GO:0030318) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 2.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 1.8 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.0 | GO:2000543 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) positive regulation of gastrulation(GO:2000543) |
| 0.0 | 4.4 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.9 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 2.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 22.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 2.4 | 14.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 2.1 | 10.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 1.4 | 5.6 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 1.3 | 3.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.1 | 11.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.0 | 9.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.8 | 5.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.7 | 6.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.6 | 8.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.6 | 1.8 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.6 | 3.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.4 | 4.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.3 | 1.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 5.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.3 | 8.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 3.0 | GO:0072357 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 4.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 1.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 6.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.2 | 1.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 6.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 4.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 2.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 7.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 11.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.4 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 5.8 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 3.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 5.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 9.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.8 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 5.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 6.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 3.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 13.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 11.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 1.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.0 | GO:0097546 | ciliary tip(GO:0097542) ciliary base(GO:0097546) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 2.2 | 22.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 1.9 | 7.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.6 | 6.5 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 1.6 | 6.2 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.3 | 4.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 1.0 | 7.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.9 | 6.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.9 | 11.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.7 | 14.3 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.7 | 7.6 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.7 | 7.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.7 | 3.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 5.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.5 | 10.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.5 | 3.0 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.5 | 1.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.4 | 9.1 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.3 | 4.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 1.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 2.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.3 | 5.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 3.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 5.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 8.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 1.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.9 | GO:0033265 | choline binding(GO:0033265) |
| 0.2 | 5.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 6.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 7.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 6.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 4.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 5.8 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 2.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 3.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 6.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.8 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.1 | 3.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 2.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 4.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 4.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 23.3 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 4.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.9 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 5.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 6.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 1.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 1.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.0 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 3.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 19.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 12.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 6.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 9.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 6.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 4.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 8.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 10.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 13.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 9.9 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 4.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 5.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 11.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.5 | 8.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 9.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.5 | 7.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 7.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 20.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 6.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 16.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 2.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 9.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 10.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 6.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 4.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 12.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |