Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
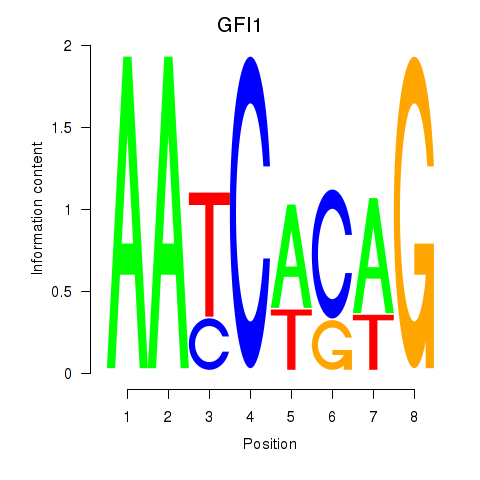
Results for GFI1
Z-value: 0.74
Transcription factors associated with GFI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GFI1
|
ENSG00000162676.7 | growth factor independent 1 transcriptional repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GFI1 | hg19_v2_chr1_-_92951607_92951661, hg19_v2_chr1_-_92949505_92949543, hg19_v2_chr1_-_92952433_92952489 | 0.14 | 3.6e-02 | Click! |
Activity profile of GFI1 motif
Sorted Z-values of GFI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_26694979 | 11.66 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr17_+_1665345 | 11.48 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_-_26695013 | 10.84 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr6_+_31982539 | 9.61 |
ENST00000435363.2
ENST00000425700.2 |
C4B
|
complement component 4B (Chido blood group) |
| chr6_+_31949801 | 9.60 |
ENST00000428956.2
ENST00000498271.1 |
C4A
|
complement component 4A (Rodgers blood group) |
| chr11_-_117748138 | 9.42 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr6_+_31795506 | 7.51 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr17_+_1665253 | 7.43 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr14_-_22005062 | 7.21 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr14_-_22005343 | 7.20 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr2_-_89157161 | 6.35 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr4_-_155533787 | 5.49 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr5_+_66254698 | 5.48 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_17721920 | 5.38 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr6_-_52859046 | 5.02 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr5_-_35230434 | 4.87 |
ENST00000504500.1
|
PRLR
|
prolactin receptor |
| chr1_-_173793458 | 4.75 |
ENST00000356198.2
|
CENPL
|
centromere protein L |
| chr15_+_84115868 | 4.67 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr10_+_124221036 | 4.63 |
ENST00000368984.3
|
HTRA1
|
HtrA serine peptidase 1 |
| chr8_-_27115931 | 4.60 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr15_+_84116106 | 4.60 |
ENST00000535412.1
ENST00000324537.5 |
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr2_+_17721230 | 4.60 |
ENST00000457525.1
|
VSNL1
|
visinin-like 1 |
| chr6_+_45296391 | 4.60 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr7_+_100136811 | 4.57 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr17_-_66951474 | 4.53 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr11_+_71791849 | 4.47 |
ENST00000423494.2
ENST00000539587.1 ENST00000538478.1 ENST00000324866.7 ENST00000439209.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr7_+_98246588 | 4.40 |
ENST00000265634.3
|
NPTX2
|
neuronal pentraxin II |
| chr1_-_153348067 | 4.34 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr6_+_45296048 | 4.30 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr19_+_50353944 | 4.29 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr14_-_93214915 | 4.27 |
ENST00000553918.1
ENST00000555699.1 ENST00000553802.1 ENST00000554397.1 ENST00000554919.1 ENST00000554080.1 ENST00000553371.1 |
LGMN
|
legumain |
| chr11_+_117049445 | 4.19 |
ENST00000324225.4
ENST00000532960.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr19_+_47813110 | 4.18 |
ENST00000355085.3
|
C5AR1
|
complement component 5a receptor 1 |
| chr19_-_49140609 | 4.15 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr19_+_55477711 | 4.06 |
ENST00000448584.2
ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chrX_+_133507327 | 4.04 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr14_-_93214988 | 4.02 |
ENST00000557434.1
ENST00000393218.2 ENST00000334869.4 |
LGMN
|
legumain |
| chr6_-_46293378 | 4.01 |
ENST00000330430.6
|
RCAN2
|
regulator of calcineurin 2 |
| chrX_+_37639302 | 4.01 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr8_-_27115903 | 3.95 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr11_+_117049854 | 3.95 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr11_+_71900703 | 3.90 |
ENST00000393681.2
|
FOLR1
|
folate receptor 1 (adult) |
| chr17_-_56606705 | 3.86 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr5_+_36608422 | 3.77 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr11_+_71791693 | 3.77 |
ENST00000289488.2
ENST00000447974.1 |
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr17_-_56606664 | 3.76 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr19_-_19302931 | 3.75 |
ENST00000444486.3
ENST00000514819.3 ENST00000585679.1 ENST00000162023.5 |
MEF2BNB-MEF2B
MEF2BNB
MEF2B
|
MEF2BNB-MEF2B readthrough MEF2B neighbor myocyte enhancer factor 2B |
| chr5_-_88179302 | 3.75 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr3_-_187388173 | 3.71 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr4_-_89744314 | 3.67 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr3_-_114343039 | 3.66 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_+_71900572 | 3.65 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr17_-_56606639 | 3.64 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chr4_-_681114 | 3.61 |
ENST00000503156.1
|
MFSD7
|
major facilitator superfamily domain containing 7 |
| chr1_-_173793246 | 3.61 |
ENST00000345664.6
ENST00000367710.3 |
CENPL
|
centromere protein L |
| chr2_+_148602058 | 3.60 |
ENST00000241416.7
ENST00000535787.1 ENST00000404590.1 |
ACVR2A
|
activin A receptor, type IIA |
| chr17_+_3539998 | 3.55 |
ENST00000452111.1
ENST00000574776.1 ENST00000441220.2 ENST00000414524.2 |
CTNS
|
cystinosin, lysosomal cystine transporter |
| chr19_-_44123734 | 3.51 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr17_-_7017968 | 3.50 |
ENST00000355035.5
|
ASGR2
|
asialoglycoprotein receptor 2 |
| chr13_-_67802549 | 3.42 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr19_+_41103063 | 3.39 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr17_-_7018128 | 3.38 |
ENST00000380952.2
ENST00000254850.7 |
ASGR2
|
asialoglycoprotein receptor 2 |
| chr11_-_82746587 | 3.38 |
ENST00000528379.1
ENST00000534103.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr2_-_152830479 | 3.36 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr12_+_8276495 | 3.35 |
ENST00000546339.1
|
CLEC4A
|
C-type lectin domain family 4, member A |
| chr4_-_89744365 | 3.33 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr6_+_31553978 | 3.31 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr19_-_49140692 | 3.31 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr8_-_119964434 | 3.30 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr6_+_41040678 | 3.28 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr19_+_55476620 | 3.27 |
ENST00000543010.1
ENST00000391721.4 ENST00000339757.7 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr14_-_23285069 | 3.27 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_-_152830441 | 3.26 |
ENST00000534999.1
ENST00000397327.2 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr6_+_31554636 | 3.26 |
ENST00000433492.1
|
LST1
|
leukocyte specific transcript 1 |
| chr22_-_32334403 | 3.24 |
ENST00000543051.1
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr3_+_40518599 | 3.23 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr1_+_163039143 | 3.19 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr15_-_23932437 | 3.18 |
ENST00000331837.4
|
NDN
|
necdin, melanoma antigen (MAGE) family member |
| chr14_-_70263979 | 3.17 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr8_+_58890917 | 3.14 |
ENST00000522992.1
|
RP11-1112C15.1
|
RP11-1112C15.1 |
| chr1_-_67142710 | 3.12 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr1_-_120311517 | 3.11 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr3_-_66024213 | 3.10 |
ENST00000483466.1
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_+_66662690 | 3.08 |
ENST00000488550.1
|
MEIS1
|
Meis homeobox 1 |
| chr1_-_230513367 | 3.08 |
ENST00000321327.2
ENST00000525115.1 |
PGBD5
|
piggyBac transposable element derived 5 |
| chr2_+_189839046 | 3.07 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr19_-_17488143 | 3.04 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chrX_-_14047996 | 3.02 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chr9_+_71939488 | 3.01 |
ENST00000455972.1
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr19_-_44124019 | 3.00 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr2_+_183943464 | 2.96 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr14_-_23285011 | 2.91 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr6_+_31553901 | 2.89 |
ENST00000418507.2
ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1
|
leukocyte specific transcript 1 |
| chr17_+_19281034 | 2.88 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr2_+_79412357 | 2.87 |
ENST00000466387.1
|
CTNNA2
|
catenin (cadherin-associated protein), alpha 2 |
| chr17_+_68071458 | 2.87 |
ENST00000589377.1
|
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr18_-_53257027 | 2.84 |
ENST00000568740.1
ENST00000564403.2 ENST00000537578.1 |
TCF4
|
transcription factor 4 |
| chr8_+_133931648 | 2.83 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr17_+_4675175 | 2.81 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr6_-_135271260 | 2.81 |
ENST00000265605.2
|
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr2_+_66662510 | 2.81 |
ENST00000272369.9
ENST00000407092.2 |
MEIS1
|
Meis homeobox 1 |
| chr12_+_81110684 | 2.79 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chrX_-_102319092 | 2.78 |
ENST00000372728.3
|
BEX1
|
brain expressed, X-linked 1 |
| chr11_-_65548265 | 2.76 |
ENST00000532090.2
|
AP5B1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_+_164528866 | 2.74 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_173793777 | 2.73 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr17_+_76311791 | 2.73 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr3_+_46412345 | 2.72 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr2_+_157292859 | 2.71 |
ENST00000438166.2
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr6_-_135271219 | 2.71 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chrX_-_55020511 | 2.65 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr11_-_71791518 | 2.65 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr12_+_113659234 | 2.64 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr3_+_186435137 | 2.64 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr5_+_178368186 | 2.63 |
ENST00000320129.3
ENST00000519564.1 |
ZNF454
|
zinc finger protein 454 |
| chr10_-_100995540 | 2.62 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr10_-_100995603 | 2.60 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr11_+_2323349 | 2.60 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chrX_+_48681768 | 2.60 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr1_+_196621002 | 2.58 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr10_-_82049424 | 2.57 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr4_+_56814968 | 2.56 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr2_+_1418154 | 2.55 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr4_+_84377115 | 2.54 |
ENST00000295491.4
ENST00000507019.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr11_-_104034827 | 2.49 |
ENST00000393158.2
|
PDGFD
|
platelet derived growth factor D |
| chr2_+_26915584 | 2.49 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chr9_-_79520989 | 2.46 |
ENST00000376713.3
ENST00000376718.3 ENST00000428286.1 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr21_-_36421535 | 2.46 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chrX_+_84499081 | 2.42 |
ENST00000276123.3
|
ZNF711
|
zinc finger protein 711 |
| chr7_-_140482926 | 2.38 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr17_-_56595196 | 2.38 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr5_+_140588269 | 2.37 |
ENST00000541609.1
ENST00000239450.2 |
PCDHB12
|
protocadherin beta 12 |
| chr19_-_18391708 | 2.37 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr17_-_62340581 | 2.36 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr4_-_89744457 | 2.36 |
ENST00000395002.2
|
FAM13A
|
family with sequence similarity 13, member A |
| chr15_+_92397051 | 2.36 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chrX_+_138612889 | 2.35 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr11_-_72432950 | 2.34 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr5_+_140729649 | 2.34 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr13_+_31191920 | 2.33 |
ENST00000255304.4
|
USPL1
|
ubiquitin specific peptidase like 1 |
| chr17_+_68071389 | 2.33 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_114477962 | 2.32 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_9143786 | 2.32 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr11_-_4629388 | 2.31 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr6_+_31554612 | 2.31 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chrX_+_84499038 | 2.30 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr14_-_106069247 | 2.30 |
ENST00000479229.1
|
RP11-731F5.1
|
RP11-731F5.1 |
| chrY_+_16168097 | 2.29 |
ENST00000250823.4
|
VCY1B
|
variable charge, Y-linked 1B |
| chr20_-_23066953 | 2.28 |
ENST00000246006.4
|
CD93
|
CD93 molecule |
| chr3_+_186435065 | 2.28 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr19_-_15529790 | 2.26 |
ENST00000596195.1
ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L
|
A kinase (PRKA) anchor protein 8-like |
| chr7_+_86273218 | 2.26 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chrX_+_100353153 | 2.25 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr6_+_6588316 | 2.24 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr6_+_31554456 | 2.24 |
ENST00000339530.4
|
LST1
|
leukocyte specific transcript 1 |
| chr11_-_71791726 | 2.23 |
ENST00000393695.3
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr20_-_3065362 | 2.22 |
ENST00000380293.3
|
AVP
|
arginine vasopressin |
| chr18_-_59854203 | 2.21 |
ENST00000589339.1
ENST00000357637.5 ENST00000585458.1 ENST00000400334.3 ENST00000587134.1 ENST00000585923.1 ENST00000590765.1 ENST00000589720.1 ENST00000588571.1 ENST00000585344.1 |
PIGN
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr1_+_176432298 | 2.21 |
ENST00000367661.3
ENST00000367662.3 |
PAPPA2
|
pappalysin 2 |
| chr7_-_27187393 | 2.20 |
ENST00000222728.3
|
HOXA6
|
homeobox A6 |
| chr3_-_38835501 | 2.19 |
ENST00000449082.2
|
SCN10A
|
sodium channel, voltage-gated, type X, alpha subunit |
| chr1_+_156338993 | 2.19 |
ENST00000368249.1
ENST00000368246.2 ENST00000537040.1 ENST00000400992.2 ENST00000255013.3 ENST00000451864.2 |
RHBG
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr2_+_85360499 | 2.19 |
ENST00000282111.3
|
TCF7L1
|
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr17_+_3539744 | 2.19 |
ENST00000046640.3
ENST00000381870.3 |
CTNS
|
cystinosin, lysosomal cystine transporter |
| chr5_-_142000883 | 2.18 |
ENST00000359370.6
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr5_+_140261703 | 2.17 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr3_-_114477787 | 2.17 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_+_56732658 | 2.15 |
ENST00000228534.4
|
IL23A
|
interleukin 23, alpha subunit p19 |
| chr11_+_61717279 | 2.14 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr18_-_53177984 | 2.14 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr4_-_144940477 | 2.12 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr11_+_61717336 | 2.11 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr4_-_145061788 | 2.10 |
ENST00000512064.1
ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA
GYPB
|
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chrX_+_84498989 | 2.09 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr7_-_87104963 | 2.06 |
ENST00000359206.3
ENST00000358400.3 ENST00000265723.4 |
ABCB4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr17_-_8093471 | 2.05 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr18_-_78005231 | 2.05 |
ENST00000470488.2
ENST00000353265.3 |
PARD6G
|
par-6 family cell polarity regulator gamma |
| chr9_+_42717234 | 2.04 |
ENST00000377590.1
|
FOXD4L2
|
forkhead box D4-like 2 |
| chr2_+_233390863 | 2.04 |
ENST00000449596.1
ENST00000543200.1 |
CHRND
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr12_+_81101277 | 2.02 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr16_+_31366455 | 2.02 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chrX_+_12885183 | 2.00 |
ENST00000380659.3
|
TLR7
|
toll-like receptor 7 |
| chr5_+_147763539 | 1.99 |
ENST00000296701.6
ENST00000394370.3 |
FBXO38
|
F-box protein 38 |
| chr14_+_103995546 | 1.99 |
ENST00000299202.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae) |
| chr5_+_147763498 | 1.98 |
ENST00000340253.5
|
FBXO38
|
F-box protein 38 |
| chr16_+_67360712 | 1.95 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr4_+_106631966 | 1.95 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr16_+_82090028 | 1.94 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr21_-_36421401 | 1.94 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr16_-_90038866 | 1.93 |
ENST00000314994.3
|
CENPBD1
|
CENPB DNA-binding domains containing 1 |
| chr9_-_35115836 | 1.91 |
ENST00000378566.1
ENST00000378554.2 ENST00000322813.5 |
FAM214B
|
family with sequence similarity 214, member B |
| chr11_+_6280927 | 1.91 |
ENST00000334619.2
|
CCKBR
|
cholecystokinin B receptor |
| chr7_-_115670792 | 1.91 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chrX_-_48056199 | 1.90 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr19_+_55417530 | 1.89 |
ENST00000350790.5
ENST00000338835.5 ENST00000357397.5 |
NCR1
|
natural cytotoxicity triggering receptor 1 |
| chr7_-_120498357 | 1.88 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr19_-_16045619 | 1.88 |
ENST00000402119.4
|
CYP4F11
|
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr12_+_110011571 | 1.86 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr12_+_8276224 | 1.85 |
ENST00000229332.5
|
CLEC4A
|
C-type lectin domain family 4, member A |
| chr19_-_15311713 | 1.84 |
ENST00000601011.1
ENST00000263388.2 |
NOTCH3
|
notch 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of GFI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 18.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 2.8 | 8.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 2.5 | 7.5 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 2.3 | 11.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 1.9 | 7.5 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 1.6 | 4.9 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.5 | 4.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 1.5 | 19.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 1.4 | 4.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 1.4 | 4.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 1.3 | 11.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.3 | 1.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.3 | 7.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 1.2 | 3.5 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 1.1 | 6.8 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 1.1 | 8.9 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 1.1 | 4.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 1.1 | 4.2 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 1.0 | 3.0 | GO:0097254 | renal tubular secretion(GO:0097254) |
| 1.0 | 5.7 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.9 | 3.7 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.9 | 4.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.9 | 3.7 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.9 | 3.6 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.9 | 4.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.9 | 9.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.9 | 2.6 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.8 | 2.5 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.8 | 4.0 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.8 | 3.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.7 | 2.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.7 | 2.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.7 | 3.5 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.7 | 2.1 | GO:0097327 | response to antineoplastic agent(GO:0097327) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.7 | 2.0 | GO:1902866 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.7 | 2.0 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.6 | 2.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.6 | 1.3 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.6 | 1.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.6 | 3.1 | GO:0070997 | neuron death(GO:0070997) |
| 0.6 | 1.8 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.6 | 2.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.6 | 1.7 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.5 | 1.6 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 3.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 | 1.6 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.5 | 1.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.5 | 5.5 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 2.9 | GO:0033121 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.5 | 5.8 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.5 | 1.4 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.5 | 8.2 | GO:0042424 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.5 | 4.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.5 | 2.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.4 | 1.3 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.4 | 2.7 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.4 | 4.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.4 | 2.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 2.2 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.4 | 1.2 | GO:0043132 | NAD transport(GO:0043132) |
| 0.4 | 2.0 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.4 | 0.8 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.4 | 2.7 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 3.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 | 12.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.4 | 3.3 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.4 | 1.8 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 6.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.4 | 1.1 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.4 | 1.4 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.3 | 1.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.3 | 5.0 | GO:0002223 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.3 | 5.9 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.3 | 2.8 | GO:0015705 | iodide transport(GO:0015705) |
| 0.3 | 0.9 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.3 | 1.8 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.3 | 0.9 | GO:0035963 | cellular response to interleukin-13(GO:0035963) |
| 0.3 | 2.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 2.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.3 | 1.5 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 2.9 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.3 | 0.9 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.3 | 1.7 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.3 | 1.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 1.4 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.3 | 1.8 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.3 | 3.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 2.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 9.5 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.2 | 2.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.2 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 1.7 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) cornea development in camera-type eye(GO:0061303) |
| 0.2 | 0.7 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 0.7 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 4.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 8.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.2 | 6.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 2.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.4 | GO:0009749 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.2 | 6.5 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.2 | 5.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.2 | 0.6 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.2 | 2.1 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.2 | 3.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.2 | 3.0 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.2 | 0.9 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.9 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 1.8 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.2 | 4.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 7.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 1.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 2.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 5.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 2.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 5.1 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.2 | 1.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.2 | 0.8 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 1.7 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.2 | 2.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.6 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 3.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 5.7 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 7.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 1.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 6.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.5 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 3.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.8 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.6 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 4.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 2.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 6.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.7 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 2.5 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 2.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 3.2 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 8.9 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.1 | 0.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 2.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 6.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.8 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 2.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 13.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.7 | GO:1901898 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 3.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 5.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.1 | 0.8 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 5.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 2.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.5 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.1 | 1.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.1 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 3.5 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 1.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.4 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 1.6 | GO:0006869 | lipid transport(GO:0006869) lipid localization(GO:0010876) |
| 0.1 | 1.1 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.1 | 2.9 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 1.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.2 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.1 | 0.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 1.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 2.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.2 | GO:0050900 | leukocyte migration(GO:0050900) |
| 0.1 | 0.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 2.2 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 0.9 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.9 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 1.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 1.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.4 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 2.3 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 1.0 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 3.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0072278 | thyroid-stimulating hormone secretion(GO:0070460) metanephric comma-shaped body morphogenesis(GO:0072278) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 1.1 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 1.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 2.2 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.6 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.5 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 2.2 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.9 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 1.4 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.7 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.0 | 0.3 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.7 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.0 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 2.4 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 4.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 2.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 2.6 | GO:0007417 | central nervous system development(GO:0007417) |
| 0.0 | 0.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.3 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 2.2 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 1.7 | GO:0006665 | sphingolipid metabolic process(GO:0006665) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 11.7 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.6 | 4.9 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 1.6 | 18.9 | GO:0043203 | axon hillock(GO:0043203) |
| 1.2 | 8.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 1.0 | 3.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.9 | 3.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.8 | 11.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.7 | 3.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.7 | 2.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.6 | 3.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.5 | 7.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 5.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 1.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 1.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 3.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.4 | 1.8 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 0.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 10.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 5.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 0.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.3 | 1.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.3 | 3.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 1.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 2.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.3 | 3.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 4.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 2.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 11.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 9.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 0.7 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.2 | 7.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 1.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 3.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 2.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 6.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 6.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 2.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 28.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 4.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 3.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 1.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 13.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 9.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 26.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 4.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 5.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 4.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 16.4 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 13.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 2.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 3.2 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 5.9 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 39.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 12.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.2 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.5 | GO:0098794 | postsynapse(GO:0098794) |
| 0.0 | 4.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.6 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.5 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 1.9 | 7.5 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.6 | 9.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.5 | 7.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 1.5 | 11.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.4 | 4.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 1.3 | 5.2 | GO:0030305 | heparanase activity(GO:0030305) |
| 1.2 | 3.5 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 1.2 | 8.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 1.2 | 3.5 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 1.0 | 5.7 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.9 | 4.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.9 | 3.5 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.9 | 2.6 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.9 | 5.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.9 | 6.9 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.8 | 6.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.8 | 11.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.8 | 2.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.7 | 2.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.7 | 7.5 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.7 | 4.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 2.0 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.6 | 2.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.6 | 3.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.6 | 6.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.5 | 1.6 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.5 | 3.8 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.5 | 2.7 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.5 | 1.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.5 | 1.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 5.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.5 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 2.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.4 | 2.2 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.4 | 1.3 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) azole transmembrane transporter activity(GO:1901474) |
| 0.4 | 2.5 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.4 | 7.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.4 | 5.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 5.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.5 | GO:0015265 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.4 | 4.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 9.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 1.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 1.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.3 | 1.0 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.3 | 1.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 3.0 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.3 | 2.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.3 | 2.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.3 | 5.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 2.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 3.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 1.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 5.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 0.7 | GO:0086079 | gap junction channel activity involved in atrial cardiac muscle cell-AV node cell electrical coupling(GO:0086076) gap junction channel activity involved in bundle of His cell-Purkinje myocyte electrical coupling(GO:0086078) gap junction channel activity involved in Purkinje myocyte-ventricular cardiac muscle cell electrical coupling(GO:0086079) |
| 0.2 | 1.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.7 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.2 | 2.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 1.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 3.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 3.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 4.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 2.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 2.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 4.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 0.6 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.2 | 0.6 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 2.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 3.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.2 | 6.7 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 18.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 7.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.2 | 2.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 3.4 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 7.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 2.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.4 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 4.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 2.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 1.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.4 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 1.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 6.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 2.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 2.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 2.7 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 3.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 1.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 3.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.3 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 1.0 | GO:0005372 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.1 | 0.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.2 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.1 | 2.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 3.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.1 | 0.2 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 35.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.7 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 2.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.6 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 1.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.4 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 8.4 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 7.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 13.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 5.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 1.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 12.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 13.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 8.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 9.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 9.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 31.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 3.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 1.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.5 | 2.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.5 | 9.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 5.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 7.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.4 | 2.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 7.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 9.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.3 | 2.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 13.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 4.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 3.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 8.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 4.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.2 | 5.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 2.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 1.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 2.9 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.9 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 2.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 10.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 6.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 4.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 2.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 5.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 5.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 3.0 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 15.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 3.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 3.8 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 3.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 6.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 11.5 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 2.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 3.5 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 5.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.9 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.9 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.8 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |