Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
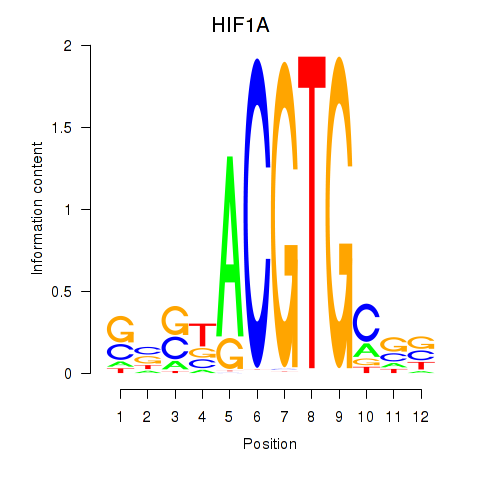
Results for HIF1A
Z-value: 0.82
Transcription factors associated with HIF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIF1A
|
ENSG00000100644.12 | hypoxia inducible factor 1 subunit alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIF1A | hg19_v2_chr14_+_62162258_62162269 | 0.61 | 7.8e-24 | Click! |
Activity profile of HIF1A motif
Sorted Z-values of HIF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_8939265 | 51.74 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr11_-_64014379 | 32.94 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr8_+_98656336 | 27.61 |
ENST00000336273.3
|
MTDH
|
metadherin |
| chr3_-_195808952 | 25.91 |
ENST00000540528.1
ENST00000392396.3 ENST00000535031.1 ENST00000420415.1 |
TFRC
|
transferrin receptor |
| chr1_-_8938736 | 25.30 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr3_-_195808980 | 25.15 |
ENST00000360110.4
|
TFRC
|
transferrin receptor |
| chr11_-_14665163 | 24.64 |
ENST00000418988.2
|
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1 |
| chr12_+_131356582 | 24.23 |
ENST00000448750.3
ENST00000541630.1 ENST00000392369.2 ENST00000254675.3 ENST00000535090.1 ENST00000392367.3 |
RAN
|
RAN, member RAS oncogene family |
| chr3_-_145878954 | 23.21 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr12_+_6977258 | 22.36 |
ENST00000488464.2
ENST00000535434.1 ENST00000493987.1 |
TPI1
|
triosephosphate isomerase 1 |
| chrX_+_77359671 | 21.54 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chrX_+_77359726 | 21.17 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr14_+_56046990 | 19.04 |
ENST00000438792.2
ENST00000395314.3 ENST00000395308.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_+_49962495 | 18.83 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chrX_+_69509927 | 18.49 |
ENST00000374403.3
|
KIF4A
|
kinesin family member 4A |
| chr5_+_138609441 | 18.22 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr1_-_159894319 | 16.96 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr14_+_64970662 | 16.75 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr1_-_26233423 | 16.68 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr15_+_74218787 | 16.66 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr12_-_21810726 | 16.62 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr12_-_21810765 | 16.17 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr14_+_56046914 | 15.89 |
ENST00000413890.2
ENST00000395309.3 ENST00000554567.1 ENST00000555498.1 |
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_-_81811312 | 15.61 |
ENST00000429644.2
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr12_-_58146128 | 15.00 |
ENST00000551800.1
ENST00000549606.1 ENST00000257904.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr8_+_55047763 | 14.71 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr15_+_52311398 | 14.54 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr16_-_81129951 | 14.40 |
ENST00000315467.3
|
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr4_-_104119528 | 14.15 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr12_-_58146048 | 13.99 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr8_+_97274119 | 13.79 |
ENST00000455950.2
|
PTDSS1
|
phosphatidylserine synthase 1 |
| chr16_-_81129845 | 13.73 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr8_-_54755459 | 13.73 |
ENST00000524234.1
ENST00000521275.1 ENST00000396774.2 |
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr14_-_64010046 | 13.71 |
ENST00000337537.3
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr11_-_122931881 | 13.70 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr11_-_64013663 | 13.55 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr14_-_54908043 | 13.25 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr17_-_57184260 | 13.16 |
ENST00000376149.3
ENST00000393066.3 |
TRIM37
|
tripartite motif containing 37 |
| chr17_-_5342380 | 13.15 |
ENST00000225698.4
|
C1QBP
|
complement component 1, q subcomponent binding protein |
| chr8_+_98656693 | 13.11 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr2_+_216176540 | 13.04 |
ENST00000236959.9
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr17_-_57184064 | 13.03 |
ENST00000262294.7
|
TRIM37
|
tripartite motif containing 37 |
| chr12_+_6643676 | 12.94 |
ENST00000396856.1
ENST00000396861.1 |
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr16_+_447209 | 12.48 |
ENST00000382940.4
ENST00000219479.2 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr8_-_54755789 | 12.46 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr1_-_6269448 | 12.46 |
ENST00000465335.1
|
RPL22
|
ribosomal protein L22 |
| chr2_+_187350883 | 12.33 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr1_-_26232951 | 12.10 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr2_+_216176761 | 11.94 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr1_-_6269304 | 11.90 |
ENST00000471204.1
|
RPL22
|
ribosomal protein L22 |
| chr3_+_25831567 | 11.86 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr13_-_31038370 | 11.86 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr8_-_97273807 | 11.63 |
ENST00000517720.1
ENST00000287025.3 ENST00000523821.1 |
MTERFD1
|
MTERF domain containing 1 |
| chr8_-_102218292 | 11.59 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr12_+_72148614 | 11.58 |
ENST00000261263.3
|
RAB21
|
RAB21, member RAS oncogene family |
| chr3_+_52719936 | 11.29 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr11_-_47664072 | 10.82 |
ENST00000542981.1
ENST00000530428.1 ENST00000302503.3 |
MTCH2
|
mitochondrial carrier 2 |
| chr6_-_153304148 | 10.53 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr6_-_114292449 | 10.43 |
ENST00000519065.1
|
HDAC2
|
histone deacetylase 2 |
| chr2_+_187350973 | 10.39 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr4_-_24586140 | 10.32 |
ENST00000336812.4
|
DHX15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr4_-_39529049 | 10.25 |
ENST00000501493.2
ENST00000509391.1 ENST00000507089.1 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr12_-_50677255 | 10.21 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr22_-_30987849 | 10.20 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr1_-_6259641 | 10.17 |
ENST00000234875.4
|
RPL22
|
ribosomal protein L22 |
| chr17_+_73201754 | 10.11 |
ENST00000583569.1
ENST00000245544.4 ENST00000579324.1 ENST00000541827.1 ENST00000579298.1 ENST00000447371.2 |
NUP85
|
nucleoporin 85kDa |
| chr20_+_56964253 | 9.81 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_-_6259613 | 9.61 |
ENST00000465387.1
|
RPL22
|
ribosomal protein L22 |
| chr2_+_242254507 | 9.56 |
ENST00000391973.2
|
SEPT2
|
septin 2 |
| chr10_+_76969909 | 9.54 |
ENST00000298468.5
ENST00000543351.1 |
VDAC2
|
voltage-dependent anion channel 2 |
| chr3_+_184080790 | 9.50 |
ENST00000430783.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr8_-_117768023 | 9.46 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr13_+_27998681 | 9.46 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr8_+_26240414 | 9.40 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr6_-_114292284 | 9.37 |
ENST00000520895.1
ENST00000521163.1 ENST00000524334.1 ENST00000368632.2 ENST00000398283.2 |
HDAC2
|
histone deacetylase 2 |
| chr1_+_112162381 | 9.02 |
ENST00000433097.1
ENST00000369709.3 ENST00000436150.2 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr8_+_109455845 | 8.83 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr1_-_43833628 | 8.81 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr10_+_75910960 | 8.76 |
ENST00000539909.1
ENST00000286621.2 |
ADK
|
adenosine kinase |
| chrX_-_102941596 | 8.58 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr9_+_100745615 | 8.47 |
ENST00000339399.4
|
ANP32B
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr1_+_193091080 | 8.44 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr19_-_5719860 | 8.42 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr22_+_24236191 | 8.39 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr1_+_230202936 | 8.33 |
ENST00000366672.4
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr15_+_89182178 | 8.31 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_-_49318715 | 8.23 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr22_-_30987837 | 8.17 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr16_+_447226 | 8.17 |
ENST00000433358.1
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr5_+_134094461 | 7.86 |
ENST00000452510.2
ENST00000354283.4 |
DDX46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr7_+_128095945 | 7.83 |
ENST00000257696.4
|
HILPDA
|
hypoxia inducible lipid droplet-associated |
| chr12_+_7023491 | 7.80 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr13_+_25875785 | 7.66 |
ENST00000381747.3
|
NUPL1
|
nucleoporin like 1 |
| chrX_+_100663243 | 7.53 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr19_+_50180409 | 7.39 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr5_-_131563501 | 7.30 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr15_+_89181974 | 7.21 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_+_7023735 | 7.20 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr1_-_235324530 | 7.18 |
ENST00000447801.1
ENST00000366606.3 ENST00000429912.1 |
RBM34
|
RNA binding motif protein 34 |
| chr1_-_26232522 | 7.18 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr10_-_16859361 | 7.13 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr19_-_1568057 | 7.09 |
ENST00000402693.4
ENST00000388824.6 |
MEX3D
|
mex-3 RNA binding family member D |
| chr10_+_76970509 | 7.00 |
ENST00000332211.6
ENST00000535553.1 ENST00000313132.4 |
VDAC2
|
voltage-dependent anion channel 2 |
| chr7_-_95951334 | 6.97 |
ENST00000265631.5
|
SLC25A13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr4_-_39529180 | 6.95 |
ENST00000515021.1
ENST00000510490.1 ENST00000316423.6 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr7_-_95951432 | 6.84 |
ENST00000416240.2
|
SLC25A13
|
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr3_-_58419537 | 6.84 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr19_+_50180507 | 6.78 |
ENST00000454376.2
ENST00000524771.1 |
PRMT1
|
protein arginine methyltransferase 1 |
| chr9_+_133569108 | 6.66 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr2_+_27435734 | 6.64 |
ENST00000419744.1
|
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr12_+_110906169 | 6.59 |
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A |
| chr10_-_16859442 | 6.48 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr1_-_32801825 | 6.46 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr17_+_45608430 | 6.38 |
ENST00000322157.4
|
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr19_-_5720123 | 6.27 |
ENST00000587365.1
ENST00000585374.1 ENST00000593119.1 |
LONP1
|
lon peptidase 1, mitochondrial |
| chr20_+_48552908 | 6.22 |
ENST00000244061.2
|
RNF114
|
ring finger protein 114 |
| chr1_+_100436065 | 6.14 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr5_+_70883117 | 6.13 |
ENST00000340941.6
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr9_+_115983808 | 6.03 |
ENST00000374210.6
ENST00000374212.4 |
SLC31A1
|
solute carrier family 31 (copper transporter), member 1 |
| chr11_+_73498898 | 5.96 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr7_+_26331541 | 5.84 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr1_-_151319710 | 5.82 |
ENST00000290524.4
ENST00000437327.1 ENST00000452513.2 ENST00000368870.2 ENST00000452671.2 |
RFX5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr4_+_17578815 | 5.78 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr5_+_61602055 | 5.66 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr12_+_28343353 | 5.60 |
ENST00000539107.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr16_+_3070356 | 5.57 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr22_+_41347363 | 5.55 |
ENST00000216225.8
|
RBX1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr1_+_231376941 | 5.52 |
ENST00000436239.1
ENST00000366647.4 ENST00000366646.3 ENST00000416000.1 |
GNPAT
|
glyceronephosphate O-acyltransferase |
| chr1_-_167522982 | 5.50 |
ENST00000370509.4
|
CREG1
|
cellular repressor of E1A-stimulated genes 1 |
| chrX_-_57147748 | 5.49 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr1_+_214454492 | 5.46 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr11_+_60609537 | 5.40 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr16_+_3070313 | 5.29 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr2_+_242254679 | 5.27 |
ENST00000428282.1
ENST00000360051.3 |
SEPT2
|
septin 2 |
| chr22_+_35653445 | 5.26 |
ENST00000420166.1
ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4
|
HMG box domain containing 4 |
| chr8_-_30515693 | 5.25 |
ENST00000355904.4
|
GTF2E2
|
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr15_+_89182156 | 5.24 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr6_-_153304697 | 5.16 |
ENST00000367241.3
|
FBXO5
|
F-box protein 5 |
| chr2_+_88991162 | 5.14 |
ENST00000283646.4
|
RPIA
|
ribose 5-phosphate isomerase A |
| chr6_+_64281906 | 5.13 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr22_-_36424458 | 5.10 |
ENST00000438146.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr3_-_149688896 | 5.06 |
ENST00000239940.7
|
PFN2
|
profilin 2 |
| chr3_+_133292851 | 5.03 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr6_+_44191290 | 5.00 |
ENST00000371755.3
ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr7_-_102985035 | 4.81 |
ENST00000426036.2
ENST00000249270.7 ENST00000454277.1 ENST00000412522.1 |
DNAJC2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr9_-_123605177 | 4.79 |
ENST00000373904.5
ENST00000210313.3 |
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr1_-_51425772 | 4.72 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr17_-_8534031 | 4.54 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr5_+_126112794 | 4.49 |
ENST00000261366.5
ENST00000395354.1 |
LMNB1
|
lamin B1 |
| chr17_+_57642886 | 4.48 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chrX_-_57164058 | 4.47 |
ENST00000374906.3
|
SPIN2A
|
spindlin family, member 2A |
| chr6_+_30539153 | 4.43 |
ENST00000326195.8
ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr17_+_66508537 | 4.40 |
ENST00000392711.1
ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr3_+_133292759 | 4.35 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr8_-_103424986 | 4.32 |
ENST00000521922.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr14_+_96968707 | 4.23 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr5_+_70883178 | 4.19 |
ENST00000323375.8
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr11_+_6624970 | 4.16 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr8_-_101734308 | 4.15 |
ENST00000519004.1
ENST00000519363.1 ENST00000520142.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr8_-_101734907 | 4.14 |
ENST00000318607.5
ENST00000521865.1 ENST00000520804.1 ENST00000522720.1 ENST00000521067.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_+_17622415 | 4.13 |
ENST00000252603.2
ENST00000600923.1 |
PGLS
|
6-phosphogluconolactonase |
| chr16_-_18812746 | 4.09 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr13_+_25670268 | 4.05 |
ENST00000281589.3
|
PABPC3
|
poly(A) binding protein, cytoplasmic 3 |
| chr6_+_71377473 | 4.02 |
ENST00000370452.3
ENST00000316999.5 ENST00000370455.3 |
SMAP1
|
small ArfGAP 1 |
| chr16_+_53164833 | 4.01 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_+_6624955 | 4.01 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr17_-_8534067 | 3.95 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr8_-_103424916 | 3.79 |
ENST00000220959.4
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr11_+_6625046 | 3.77 |
ENST00000396751.2
|
ILK
|
integrin-linked kinase |
| chr17_+_40985407 | 3.71 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr2_+_178077477 | 3.67 |
ENST00000411529.2
ENST00000435711.1 |
HNRNPA3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr10_-_96122682 | 3.65 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr1_-_246729544 | 3.52 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr7_+_96747030 | 3.44 |
ENST00000360382.4
|
ACN9
|
ACN9 homolog (S. cerevisiae) |
| chr2_+_232575128 | 3.40 |
ENST00000412128.1
|
PTMA
|
prothymosin, alpha |
| chr17_+_42148225 | 3.40 |
ENST00000591696.1
|
G6PC3
|
glucose 6 phosphatase, catalytic, 3 |
| chr8_-_95908902 | 3.27 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr3_-_50329990 | 3.27 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chrX_-_57147902 | 3.24 |
ENST00000275988.5
ENST00000434397.1 ENST00000333933.3 ENST00000374912.5 |
SPIN2B
|
spindlin family, member 2B |
| chrX_+_47077632 | 3.19 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr3_+_63898275 | 3.10 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr2_-_220118631 | 3.09 |
ENST00000248437.4
|
TUBA4A
|
tubulin, alpha 4a |
| chr19_+_1407517 | 3.04 |
ENST00000336761.6
ENST00000233078.4 |
DAZAP1
|
DAZ associated protein 1 |
| chr15_-_59665062 | 3.03 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr8_-_103425047 | 2.94 |
ENST00000520539.1
|
UBR5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr2_+_85132749 | 2.94 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr12_+_28343365 | 2.93 |
ENST00000545336.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chrX_-_41782683 | 2.92 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr10_+_93558069 | 2.89 |
ENST00000371627.4
|
TNKS2
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chr4_+_83351715 | 2.83 |
ENST00000273920.3
|
ENOPH1
|
enolase-phosphatase 1 |
| chr6_+_64282447 | 2.78 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr2_+_68384976 | 2.74 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr18_+_9913977 | 2.71 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr6_-_109702885 | 2.63 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr12_-_51422017 | 2.63 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr17_+_42148097 | 2.62 |
ENST00000269097.4
|
G6PC3
|
glucose 6 phosphatase, catalytic, 3 |
| chr7_+_75931861 | 2.62 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr14_+_96968802 | 2.53 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr8_+_67341239 | 2.51 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr17_+_80477571 | 2.46 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HIF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.0 | 77.0 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 8.7 | 26.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 7.7 | 23.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 7.5 | 22.4 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 7.2 | 36.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 6.9 | 20.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 6.2 | 25.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 5.8 | 29.0 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 5.6 | 28.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 5.2 | 15.7 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 4.9 | 14.6 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 4.6 | 13.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 4.2 | 16.7 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 4.0 | 19.8 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 3.7 | 11.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 3.5 | 24.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 3.3 | 13.2 | GO:1901165 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 3.0 | 9.0 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 3.0 | 9.0 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 3.0 | 11.9 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 2.9 | 8.8 | GO:0044209 | AMP salvage(GO:0044209) |
| 2.9 | 17.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 2.8 | 8.5 | GO:0021678 | fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 2.7 | 13.7 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 2.7 | 51.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 2.4 | 11.9 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 2.4 | 14.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 2.2 | 32.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 2.1 | 8.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 2.0 | 42.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 2.0 | 14.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) negative regulation of megakaryocyte differentiation(GO:0045653) |
| 1.8 | 5.5 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 1.8 | 18.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 1.7 | 13.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 1.7 | 20.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 1.7 | 8.4 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.6 | 4.8 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 1.6 | 11.1 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 1.6 | 9.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.5 | 8.8 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 1.4 | 2.9 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.4 | 4.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 1.4 | 10.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 1.4 | 2.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 1.2 | 18.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.2 | 12.3 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 1.2 | 11.6 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 1.1 | 10.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 1.1 | 7.9 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 1.1 | 9.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 1.0 | 5.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.0 | 6.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 1.0 | 6.0 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 1.0 | 5.0 | GO:0015862 | uridine transport(GO:0015862) |
| 1.0 | 4.8 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.9 | 8.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.9 | 3.5 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.9 | 26.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.8 | 5.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.8 | 4.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.8 | 2.4 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.8 | 6.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.8 | 11.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.7 | 8.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.7 | 8.4 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.7 | 2.6 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.6 | 7.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.6 | 8.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.6 | 2.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.6 | 1.8 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.6 | 13.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.6 | 7.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.6 | 5.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.5 | 5.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.5 | 39.4 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.5 | 2.6 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.4 | 49.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.4 | 2.2 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.4 | 5.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 1.5 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.4 | 16.5 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.4 | 4.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.4 | 1.4 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.3 | 10.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 6.6 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.3 | 4.4 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.3 | 2.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 13.2 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 1.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.3 | 28.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.3 | 8.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.3 | 8.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 0.9 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.3 | 12.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.3 | 20.7 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.3 | 3.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.3 | 10.1 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.3 | 16.7 | GO:0035904 | aorta development(GO:0035904) |
| 0.2 | 1.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.9 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 | 5.8 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 2.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 2.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.2 | 3.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) |
| 0.2 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.8 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 3.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.2 | 1.6 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 10.2 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.2 | 4.0 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 6.8 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.2 | 3.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.8 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.2 | 8.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.2 | 3.4 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.2 | 10.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 1.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 37.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 6.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 2.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 9.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 20.1 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 3.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 5.3 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 2.5 | GO:0016579 | protein deubiquitination(GO:0016579) protein modification by small protein removal(GO:0070646) |
| 0.1 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 1.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 1.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 2.6 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.6 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 3.3 | GO:0007129 | synapsis(GO:0007129) |
| 0.1 | 4.0 | GO:0016569 | covalent chromatin modification(GO:0016569) |
| 0.1 | 14.8 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 1.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 6.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 1.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 4.5 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 22.6 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 4.8 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 46.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 2.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.5 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 1.5 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.9 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 11.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.3 | 92.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 9.4 | 28.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 5.8 | 40.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 5.2 | 26.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 4.8 | 29.0 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 3.9 | 51.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 3.1 | 18.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 2.1 | 10.3 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 2.1 | 10.3 | GO:1905202 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 2.1 | 24.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 2.0 | 13.7 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.9 | 5.6 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.8 | 46.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.7 | 8.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.6 | 24.2 | GO:0090543 | Flemming body(GO:0090543) |
| 1.6 | 11.0 | GO:0097452 | GAIT complex(GO:0097452) |
| 1.6 | 12.5 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 1.3 | 29.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 1.2 | 14.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.1 | 5.3 | GO:0016589 | NURF complex(GO:0016589) |
| 1.1 | 9.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 1.0 | 8.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 1.0 | 27.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 1.0 | 5.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.8 | 8.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.8 | 3.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.8 | 9.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.8 | 14.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.7 | 16.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.7 | 10.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.6 | 12.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.6 | 3.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 9.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 2.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.5 | 20.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.5 | 4.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.5 | 12.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 20.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 25.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.4 | 44.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 13.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 18.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 22.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 8.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.3 | 2.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 4.8 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 20.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 0.7 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 5.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 4.4 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 20.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 47.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 0.9 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 2.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 19.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 3.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 5.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 6.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 5.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 21.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 5.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 17.3 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 24.7 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.1 | 21.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 8.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.0 | GO:0098791 | organelle subcompartment(GO:0031984) Golgi subcompartment(GO:0098791) |
| 0.0 | 1.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 7.3 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 2.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 2.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 5.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 75.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.0 | 51.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 15.3 | 92.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 14.2 | 42.7 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 7.7 | 23.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 7.0 | 28.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 6.9 | 20.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 4.9 | 14.6 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 4.7 | 32.8 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 4.4 | 13.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 4.2 | 25.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 4.1 | 12.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 3.5 | 14.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 2.8 | 13.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 2.8 | 11.0 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 2.7 | 24.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 2.5 | 27.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 2.4 | 14.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 2.3 | 20.7 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 2.1 | 10.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 2.0 | 11.9 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 1.9 | 20.7 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 1.8 | 16.5 | GO:0015288 | porin activity(GO:0015288) |
| 1.7 | 6.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.6 | 63.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.6 | 15.7 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.5 | 6.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 1.5 | 7.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 1.4 | 8.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 1.4 | 4.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 1.2 | 13.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.2 | 12.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.1 | 5.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.1 | 50.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 1.0 | 16.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.9 | 32.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.9 | 26.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.9 | 24.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 2.6 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.9 | 8.7 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.8 | 6.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.8 | 16.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.8 | 2.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.8 | 4.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.8 | 11.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.8 | 6.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.7 | 6.7 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.7 | 2.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.7 | 3.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.6 | 34.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.6 | 19.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.6 | 1.9 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.6 | 18.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.6 | 3.7 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.6 | 5.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.6 | 17.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.6 | 8.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.6 | 2.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.5 | 26.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.5 | 3.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 9.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 8.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 8.8 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.5 | 5.5 | GO:0016413 | palmitoyl-CoA hydrolase activity(GO:0016290) O-acetyltransferase activity(GO:0016413) |
| 0.4 | 1.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.4 | 9.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 5.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 18.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 8.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.4 | 1.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.3 | 60.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 8.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.3 | 8.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 7.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.3 | 6.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 0.8 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.3 | 5.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 4.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 5.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 4.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 7.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 4.4 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 2.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.0 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 12.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.2 | 1.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 6.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 12.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.5 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.2 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 5.8 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 2.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 2.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 10.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.0 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 9.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 5.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 0.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 6.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.3 | GO:0008135 | translation factor activity, RNA binding(GO:0008135) |
| 0.1 | 1.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 32.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 6.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 1.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 10.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 1.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 3.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 3.0 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 36.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.6 | 24.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 1.5 | 213.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.8 | 58.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.7 | 34.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.5 | 30.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 11.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 19.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.3 | 3.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 7.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 10.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 5.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 44.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 9.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 10.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 5.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 5.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 4.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 4.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 4.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 2.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 166.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 2.4 | 77.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 1.5 | 25.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 1.3 | 15.7 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 1.2 | 33.7 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 1.0 | 9.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.9 | 24.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 25.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.8 | 13.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.7 | 17.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.7 | 13.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 5.6 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.6 | 20.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.6 | 55.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.6 | 11.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 26.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.5 | 29.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.4 | 22.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.4 | 10.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 8.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 9.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 6.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.4 | 3.3 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.4 | 12.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 23.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 58.9 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.3 | 16.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.3 | 6.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 20.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 8.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 8.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 4.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 4.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 2.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.2 | 7.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 5.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 16.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 9.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 9.3 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.1 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 2.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 3.5 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 1.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |