Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
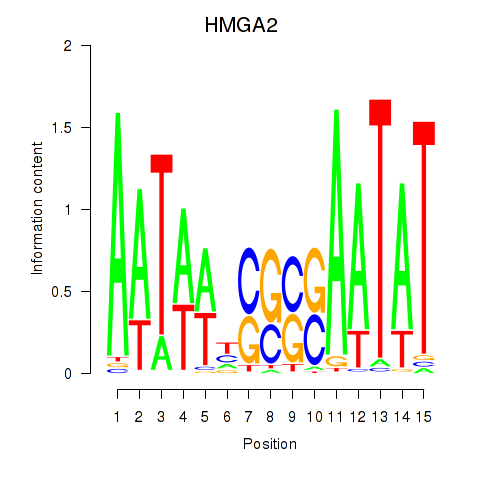
Results for HMGA2
Z-value: 0.35
Transcription factors associated with HMGA2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMGA2
|
ENSG00000149948.9 | high mobility group AT-hook 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMGA2 | hg19_v2_chr12_+_66218212_66218244 | 0.45 | 4.8e-12 | Click! |
Activity profile of HMGA2 motif
Sorted Z-values of HMGA2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_111580508 | 11.73 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr1_+_196621002 | 8.13 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr10_-_5046042 | 7.84 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr12_-_122751002 | 7.57 |
ENST00000267199.4
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae) |
| chr6_-_52705641 | 5.95 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr1_+_196621156 | 5.92 |
ENST00000359637.2
|
CFH
|
complement factor H |
| chr2_-_216257849 | 5.84 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr15_+_43985084 | 5.76 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr15_+_43885252 | 5.60 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr19_-_36297632 | 4.76 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr15_+_58702742 | 4.73 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr19_-_12267524 | 4.66 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr18_-_21017817 | 4.55 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr11_+_73661364 | 4.52 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr4_+_40058411 | 4.26 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr2_+_1417228 | 3.80 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr5_+_36608422 | 3.34 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chrX_-_7895755 | 3.24 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr9_-_115480303 | 2.83 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr1_+_55464600 | 2.68 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr14_+_99947715 | 2.66 |
ENST00000389879.5
ENST00000557441.1 ENST00000555049.1 ENST00000555842.1 |
CCNK
|
cyclin K |
| chr10_-_62332357 | 2.63 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_+_220143989 | 2.61 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chrX_-_7895479 | 2.58 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4 |
| chr2_+_172543967 | 2.52 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chrY_+_22918021 | 2.51 |
ENST00000288666.5
|
RPS4Y2
|
ribosomal protein S4, Y-linked 2 |
| chr17_-_4607335 | 2.35 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr8_-_105479270 | 2.33 |
ENST00000521573.2
ENST00000351513.2 |
DPYS
|
dihydropyrimidinase |
| chr6_+_123110302 | 2.29 |
ENST00000368440.4
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr3_-_106959424 | 2.20 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr2_+_172543919 | 2.02 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chrX_-_153523462 | 2.00 |
ENST00000361930.3
ENST00000369926.1 |
TEX28
|
testis expressed 28 |
| chr11_-_130786400 | 2.00 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr2_+_172544182 | 1.99 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr3_+_193853927 | 1.93 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr12_+_18414446 | 1.89 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr6_-_32908765 | 1.82 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr14_-_94759408 | 1.74 |
ENST00000554723.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr15_+_84904525 | 1.71 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr14_-_94759361 | 1.62 |
ENST00000393096.1
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr5_-_135290705 | 1.61 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr11_+_60260248 | 1.58 |
ENST00000526784.1
ENST00000016913.4 ENST00000537076.1 ENST00000530007.1 |
MS4A12
|
membrane-spanning 4-domains, subfamily A, member 12 |
| chr14_+_50291993 | 1.56 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr17_-_26220366 | 1.45 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr14_-_94759595 | 1.43 |
ENST00000261994.4
|
SERPINA10
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_+_172544011 | 1.39 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr18_+_11851383 | 1.38 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr2_-_183387430 | 1.37 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr19_-_38146289 | 1.21 |
ENST00000392144.1
ENST00000591444.1 ENST00000351218.2 ENST00000587809.1 |
ZFP30
|
ZFP30 zinc finger protein |
| chr11_+_69455855 | 1.17 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr19_+_44455368 | 1.16 |
ENST00000591168.1
ENST00000587682.1 ENST00000251269.5 |
ZNF221
|
zinc finger protein 221 |
| chrX_-_23926004 | 1.16 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr2_+_220144052 | 1.03 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr6_+_29426230 | 1.00 |
ENST00000442615.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr6_+_26156551 | 0.94 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr14_+_45553296 | 0.90 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr14_-_65409438 | 0.89 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr8_+_9911778 | 0.87 |
ENST00000317173.4
ENST00000441698.2 |
MSRA
|
methionine sulfoxide reductase A |
| chr15_+_28623784 | 0.68 |
ENST00000526619.2
ENST00000337838.7 ENST00000532622.2 |
GOLGA8F
|
golgin A8 family, member F |
| chr9_-_86955598 | 0.66 |
ENST00000376238.4
|
SLC28A3
|
solute carrier family 28 (concentrative nucleoside transporter), member 3 |
| chr15_+_83098710 | 0.66 |
ENST00000561062.1
ENST00000358583.3 |
GOLGA6L9
|
golgin A6 family-like 20 |
| chrX_+_117480036 | 0.52 |
ENST00000371822.5
ENST00000254029.3 ENST00000371825.3 |
WDR44
|
WD repeat domain 44 |
| chr21_+_38792602 | 0.50 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr15_+_82722225 | 0.45 |
ENST00000300515.8
|
GOLGA6L9
|
golgin A6 family-like 9 |
| chr1_+_144989309 | 0.43 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr6_-_52926539 | 0.42 |
ENST00000350082.5
ENST00000356971.3 |
ICK
|
intestinal cell (MAK-like) kinase |
| chr14_-_65409502 | 0.42 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr17_-_50236039 | 0.41 |
ENST00000451037.2
|
CA10
|
carbonic anhydrase X |
| chr12_-_8765446 | 0.39 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr19_-_11456935 | 0.31 |
ENST00000590788.1
ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr2_-_183387064 | 0.22 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_183387283 | 0.18 |
ENST00000435564.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr2_+_177134201 | 0.16 |
ENST00000452865.1
|
MTX2
|
metaxin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMGA2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 1.5 | 4.6 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 1.1 | 7.8 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.9 | 3.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.9 | 2.7 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.8 | 4.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.8 | 2.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.7 | 7.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.7 | 7.6 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.7 | 4.8 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.7 | 2.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.7 | 2.6 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.6 | 1.9 | GO:0021558 | midbrain-hindbrain boundary morphogenesis(GO:0021555) trochlear nerve development(GO:0021558) regulation of timing of neuron differentiation(GO:0060164) |
| 0.6 | 14.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.5 | 5.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.5 | 3.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 4.7 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 2.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 5.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 3.8 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.1 | 4.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.7 | GO:0015860 | pyrimidine nucleobase transport(GO:0015855) purine nucleoside transmembrane transport(GO:0015860) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 1.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 1.4 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.4 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 1.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.9 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.4 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 5.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.9 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.1 | 2.7 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 1.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.2 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 4.1 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 2.8 | GO:0010212 | response to ionizing radiation(GO:0010212) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.8 | 7.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 2.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 5.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.3 | 7.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 1.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 4.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 5.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 14.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 4.5 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 10.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 4.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.0 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 1.3 | 7.8 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 1.2 | 4.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 1.1 | 4.3 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.8 | 2.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.7 | 5.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 3.8 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.6 | 14.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.5 | 3.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 10.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.4 | 7.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 0.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.3 | 1.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 1.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.7 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.2 | 0.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.2 | 0.9 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 2.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 5.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 5.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 2.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 3.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 14.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 5.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 4.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 5.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 2.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 3.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 7.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 2.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 3.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 11.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 4.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |