Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
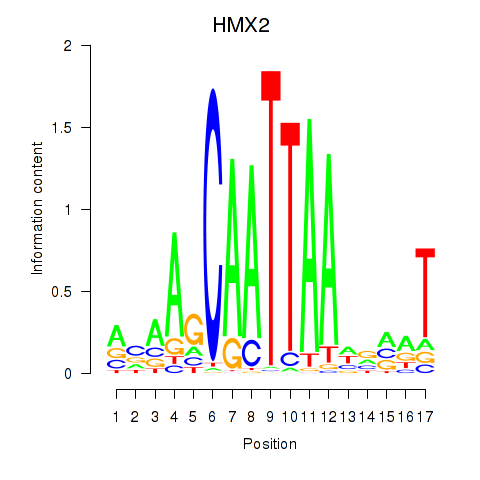
Results for HMX2
Z-value: 1.01
Transcription factors associated with HMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX2
|
ENSG00000188816.3 | H6 family homeobox 2 |
Activity profile of HMX2 motif
Sorted Z-values of HMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_95241951 | 42.86 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr12_+_104680659 | 28.63 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr2_-_151344172 | 26.17 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr7_-_87856280 | 25.91 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr7_-_87856303 | 25.17 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr10_-_95242044 | 22.76 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr5_+_167913450 | 22.70 |
ENST00000231572.3
ENST00000538719.1 |
RARS
|
arginyl-tRNA synthetase |
| chr5_+_154393260 | 20.72 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr1_+_99127225 | 20.08 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chrX_+_119737806 | 19.15 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr20_+_3776371 | 18.43 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr7_-_7680601 | 16.89 |
ENST00000396682.2
|
RPA3
|
replication protein A3, 14kDa |
| chr3_+_23847394 | 15.19 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr3_+_133502877 | 15.10 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr9_-_95055956 | 15.07 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr3_+_23847432 | 14.40 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr17_+_8191815 | 14.15 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr1_+_24019099 | 14.04 |
ENST00000443624.1
ENST00000458455.1 |
RPL11
|
ribosomal protein L11 |
| chr10_+_12238171 | 13.28 |
ENST00000378900.2
ENST00000442050.1 |
CDC123
|
cell division cycle 123 |
| chr3_-_194188956 | 13.24 |
ENST00000256031.4
ENST00000446356.1 |
ATP13A3
|
ATPase type 13A3 |
| chr10_-_121296045 | 13.22 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr2_+_118572226 | 13.21 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chrX_+_133594168 | 12.86 |
ENST00000298556.7
|
HPRT1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr11_+_60681346 | 11.77 |
ENST00000227525.3
|
TMEM109
|
transmembrane protein 109 |
| chr5_-_142814241 | 11.59 |
ENST00000504572.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_139063372 | 11.08 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr3_+_23851928 | 11.00 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr14_+_55595762 | 10.82 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr5_-_134735568 | 10.77 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr8_-_67974552 | 10.63 |
ENST00000357849.4
|
COPS5
|
COP9 signalosome subunit 5 |
| chr11_-_118972575 | 10.26 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr1_-_21377447 | 9.76 |
ENST00000374937.3
ENST00000264211.8 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr2_-_190044480 | 9.57 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr11_-_327537 | 9.22 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr12_+_64798095 | 9.05 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr17_+_48823896 | 8.92 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr13_+_27998681 | 8.85 |
ENST00000381140.4
|
GTF3A
|
general transcription factor IIIA |
| chr3_+_122785895 | 8.75 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr2_+_109237717 | 8.58 |
ENST00000409441.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_-_20251744 | 8.45 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr9_+_5890802 | 8.41 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr17_+_56769924 | 8.32 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr1_-_115259337 | 8.27 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr1_+_144151520 | 8.17 |
ENST00000369372.4
|
NBPF8
|
neuroblastoma breakpoint family, member 8 |
| chr3_-_79816965 | 7.84 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_+_114178512 | 7.66 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr13_-_103426112 | 7.58 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr3_+_179280668 | 7.50 |
ENST00000429709.2
ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A
|
actin-like 6A |
| chr6_+_12290586 | 7.42 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr14_+_55595960 | 7.40 |
ENST00000554715.1
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chr4_+_83956237 | 7.30 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr4_+_166248775 | 7.23 |
ENST00000261507.6
ENST00000507013.1 ENST00000393766.2 ENST00000504317.1 |
MSMO1
|
methylsterol monooxygenase 1 |
| chr5_+_95066823 | 7.19 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr2_+_216974020 | 7.17 |
ENST00000392132.2
ENST00000417391.1 |
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining) |
| chr4_+_83956312 | 7.17 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr2_-_55237484 | 6.89 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr17_+_1936687 | 6.83 |
ENST00000570477.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chr12_-_122985067 | 6.75 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr11_+_93517393 | 6.59 |
ENST00000251871.3
ENST00000530819.1 |
MED17
|
mediator complex subunit 17 |
| chr8_-_13134045 | 6.42 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chr15_-_101835110 | 6.36 |
ENST00000560496.1
|
SNRPA1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr13_-_103426081 | 6.19 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr10_-_126849588 | 6.10 |
ENST00000411419.2
|
CTBP2
|
C-terminal binding protein 2 |
| chr5_-_146781153 | 6.02 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr20_+_56964253 | 5.89 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_-_47239738 | 5.65 |
ENST00000413193.2
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr2_-_169104651 | 5.24 |
ENST00000355999.4
|
STK39
|
serine threonine kinase 39 |
| chr2_+_106468204 | 5.14 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr13_-_38172863 | 4.99 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr20_-_47804894 | 4.93 |
ENST00000371828.3
ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1
|
staufen double-stranded RNA binding protein 1 |
| chr3_-_52713729 | 4.82 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr5_+_119867159 | 4.76 |
ENST00000505123.1
|
PRR16
|
proline rich 16 |
| chr5_+_61602055 | 4.65 |
ENST00000381103.2
|
KIF2A
|
kinesin heavy chain member 2A |
| chr1_-_21377383 | 4.63 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr1_-_110933611 | 4.48 |
ENST00000472422.2
ENST00000437429.2 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr9_+_12693336 | 4.39 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr10_+_13203543 | 4.26 |
ENST00000378714.3
ENST00000479669.1 ENST00000484800.2 |
MCM10
|
minichromosome maintenance complex component 10 |
| chrX_-_57164058 | 4.09 |
ENST00000374906.3
|
SPIN2A
|
spindlin family, member 2A |
| chr7_-_32529973 | 4.07 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr4_+_37962018 | 3.93 |
ENST00000504686.1
|
PTTG2
|
pituitary tumor-transforming 2 |
| chr7_-_20256965 | 3.89 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr2_+_27255806 | 3.78 |
ENST00000238788.9
ENST00000404032.3 |
TMEM214
|
transmembrane protein 214 |
| chr11_+_7618413 | 3.63 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr19_+_41281416 | 3.62 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr2_-_160654745 | 3.61 |
ENST00000259053.4
ENST00000429078.2 |
CD302
|
CD302 molecule |
| chr6_+_26124373 | 3.55 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr1_-_109935819 | 3.44 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr10_+_48189612 | 3.33 |
ENST00000453919.1
|
AGAP9
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr1_-_11918988 | 3.21 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr2_-_145277569 | 3.19 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_105937068 | 3.04 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr19_+_9361606 | 3.04 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr4_-_48782259 | 2.87 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr3_+_173116225 | 2.78 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr6_-_26124138 | 2.77 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr6_-_106773491 | 2.73 |
ENST00000360666.4
|
ATG5
|
autophagy related 5 |
| chr1_-_110933663 | 2.73 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr4_-_69817481 | 2.65 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr4_-_39979576 | 2.65 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr2_+_152214098 | 2.62 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr14_-_20801427 | 2.60 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr6_+_26199737 | 2.49 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr4_-_26492076 | 2.48 |
ENST00000295589.3
|
CCKAR
|
cholecystokinin A receptor |
| chr14_+_31091511 | 2.33 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chr17_-_47755436 | 2.22 |
ENST00000505581.1
ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP
|
speckle-type POZ protein |
| chr13_+_21714913 | 2.11 |
ENST00000450573.1
ENST00000467636.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr2_-_99279928 | 1.97 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr4_-_159080806 | 1.94 |
ENST00000590648.1
|
FAM198B
|
family with sequence similarity 198, member B |
| chr1_+_186798073 | 1.94 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr7_+_141695671 | 1.92 |
ENST00000497673.1
ENST00000475668.2 |
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr7_+_141695633 | 1.92 |
ENST00000549489.2
|
MGAM
|
maltase-glucoamylase (alpha-glucosidase) |
| chr20_-_16554078 | 1.82 |
ENST00000354981.2
ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B
|
kinesin family member 16B |
| chr11_+_105480793 | 1.74 |
ENST00000282499.5
ENST00000393127.2 ENST00000527669.1 |
GRIA4
|
glutamate receptor, ionotropic, AMPA 4 |
| chr16_-_21663950 | 1.72 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr16_-_67517716 | 1.71 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr4_-_70518941 | 1.64 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr1_-_205290865 | 1.61 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr16_-_21663919 | 1.60 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr20_+_33104199 | 1.55 |
ENST00000357156.2
ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1
|
dynein, light chain, roadblock-type 1 |
| chr17_-_46671323 | 1.48 |
ENST00000239151.5
|
HOXB5
|
homeobox B5 |
| chr6_+_25652501 | 1.44 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr1_+_201979645 | 1.30 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr13_+_50570019 | 1.28 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr17_-_64225508 | 1.28 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr22_-_39268308 | 1.20 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr12_+_26164645 | 1.08 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr9_-_21385396 | 1.01 |
ENST00000380206.2
|
IFNA2
|
interferon, alpha 2 |
| chr19_+_40877583 | 0.98 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chr10_-_115613828 | 0.96 |
ENST00000361384.2
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr13_-_46756351 | 0.90 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_-_46249878 | 0.89 |
ENST00000296140.3
|
CCR1
|
chemokine (C-C motif) receptor 1 |
| chr21_-_34185944 | 0.85 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr4_-_155511887 | 0.81 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr14_-_50864006 | 0.59 |
ENST00000216378.2
|
CDKL1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr3_+_185303962 | 0.59 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr3_+_39424828 | 0.56 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr8_+_32579341 | 0.51 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr10_-_101945771 | 0.40 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr15_+_41056255 | 0.36 |
ENST00000561160.1
ENST00000559445.1 |
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr17_+_45973516 | 0.24 |
ENST00000376741.4
|
SP2
|
Sp2 transcription factor |
| chr3_-_187009798 | 0.22 |
ENST00000337774.5
|
MASP1
|
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr2_+_1417228 | 0.20 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr19_-_51340469 | 0.20 |
ENST00000326856.4
|
KLK15
|
kallikrein-related peptidase 15 |
| chr19_-_14889349 | 0.15 |
ENST00000315576.3
ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr1_-_217804377 | 0.15 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr8_+_31496809 | 0.06 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr18_+_10526008 | 0.04 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr1_-_213189168 | 0.02 |
ENST00000366962.3
ENST00000360506.2 |
ANGEL2
|
angel homolog 2 (Drosophila) |
| chr6_+_151561085 | 0.01 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_+_211342432 | 0.00 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 22.7 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 7.3 | 51.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 6.4 | 19.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 6.1 | 18.2 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 5.5 | 65.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 5.1 | 40.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 5.0 | 15.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 4.3 | 12.9 | GO:0046098 | hypoxanthine salvage(GO:0043103) guanine metabolic process(GO:0046098) |
| 3.6 | 10.8 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 3.5 | 14.1 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 3.5 | 14.0 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 3.2 | 28.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 2.6 | 7.8 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 2.5 | 7.4 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 2.4 | 9.6 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 2.4 | 7.2 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 2.1 | 10.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 1.8 | 18.4 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.7 | 5.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 1.7 | 5.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.7 | 8.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 1.6 | 4.9 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 1.6 | 25.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.4 | 11.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 1.4 | 20.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.0 | 3.8 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.9 | 2.8 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.9 | 2.7 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.9 | 5.1 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.9 | 6.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.8 | 7.6 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.8 | 2.5 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.8 | 6.9 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.7 | 13.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.7 | 14.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.7 | 6.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 6.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.6 | 16.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.5 | 1.9 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.5 | 6.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 13.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.4 | 2.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 2.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.4 | 4.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.4 | 10.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 3.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 3.0 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.3 | 3.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 8.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 13.2 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.3 | 7.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 23.8 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 11.8 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 8.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 2.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.2 | 14.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 9.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 9.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 2.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 11.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 2.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 1.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 4.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.3 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 7.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 1.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 3.9 | GO:2000816 | negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.1 | 1.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 4.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.6 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 | 4.6 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 7.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.1 | 1.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 6.4 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.6 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 2.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 6.6 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 1.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 8.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 1.7 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.9 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 1.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 12.9 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 7.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 4.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.5 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 8.1 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 1.5 | GO:0051781 | positive regulation of cell division(GO:0051781) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.3 | 51.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 3.2 | 9.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.2 | 15.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 1.9 | 7.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.8 | 37.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 1.8 | 10.8 | GO:0001740 | Barr body(GO:0001740) |
| 1.4 | 7.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 1.4 | 8.3 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.1 | 16.9 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.7 | 80.8 | GO:0005901 | caveola(GO:0005901) |
| 0.7 | 14.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.6 | 4.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.5 | 25.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.5 | 6.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.5 | 4.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.5 | 6.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.5 | 27.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.5 | 2.7 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.4 | 11.1 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.4 | 8.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.4 | 7.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.3 | 6.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.3 | 18.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 19.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 4.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 6.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 6.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 2.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 6.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 2.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 4.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 14.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 38.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 2.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 12.1 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.1 | 6.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 15.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 9.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 13.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 14.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 7.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 4.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 9.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 3.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 6.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 22.6 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 1.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 8.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 26.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 9.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 3.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 5.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 12.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.9 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.7 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.6 | GO:0098626 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 7.6 | 22.7 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 6.8 | 40.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 5.0 | 15.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 5.0 | 15.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 3.4 | 10.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 2.5 | 25.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 2.3 | 11.6 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 2.3 | 18.2 | GO:0019863 | IgE binding(GO:0019863) |
| 2.0 | 14.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.0 | 6.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 1.6 | 22.9 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 1.5 | 7.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 1.3 | 51.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 1.3 | 7.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.2 | 10.8 | GO:0000182 | rDNA binding(GO:0000182) |
| 1.0 | 8.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.0 | 3.8 | GO:0016160 | amylase activity(GO:0016160) maltose alpha-glucosidase activity(GO:0032450) |
| 0.9 | 6.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.8 | 2.5 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.8 | 6.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.7 | 22.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 7.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.7 | 3.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.7 | 14.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.6 | 6.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.5 | 13.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.5 | 2.7 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.4 | 8.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.4 | 12.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 6.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 19.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 2.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 4.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 1.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 12.9 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.2 | 9.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 2.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 5.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 7.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 13.2 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.2 | 17.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 4.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 6.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 67.6 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.2 | 7.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 1.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 7.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 4.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 41.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 7.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 6.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 12.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 10.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 1.0 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 4.4 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 5.2 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.1 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 11.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 8.9 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.4 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 7.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 9.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.1 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 4.5 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 18.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.5 | 70.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.5 | 26.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.5 | 15.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.3 | 10.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.3 | 7.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 34.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 11.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 9.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 19.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 8.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.2 | 9.4 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.2 | 8.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 10.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 7.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 10.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 6.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 3.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.0 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 4.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 40.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 1.4 | 16.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.0 | 37.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 1.0 | 25.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.9 | 18.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.8 | 28.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.7 | 7.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.7 | 8.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.7 | 18.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.5 | 13.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.5 | 12.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 7.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.3 | 3.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 8.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 23.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 4.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 7.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 11.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 5.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 6.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 9.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 14.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 10.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 4.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 2.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 14.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 1.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 6.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 6.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 3.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 6.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |