Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HMX3
Z-value: 1.08
Transcription factors associated with HMX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX3
|
ENSG00000188620.9 | H6 family homeobox 3 |
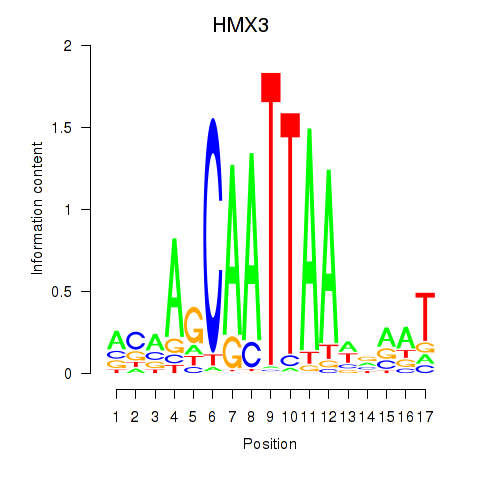
Activity profile of HMX3 motif
Sorted Z-values of HMX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.3 | 39.9 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 8.0 | 40.0 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 5.5 | 22.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 5.4 | 21.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 5.1 | 15.4 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 4.9 | 14.8 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 4.7 | 14.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 3.8 | 11.3 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 3.4 | 10.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 3.0 | 15.0 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 2.7 | 10.8 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 2.7 | 8.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 2.6 | 13.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 2.6 | 7.8 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 2.6 | 15.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 2.5 | 7.6 | GO:1903487 | regulation of lactation(GO:1903487) |
| 2.5 | 35.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 2.4 | 7.3 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 2.4 | 7.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 2.3 | 6.9 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 2.3 | 11.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 2.1 | 8.5 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 2.1 | 10.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 2.1 | 6.4 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 2.1 | 10.6 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 2.1 | 6.3 | GO:2001302 | cellular response to interleukin-13(GO:0035963) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 2.1 | 25.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 2.1 | 10.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 2.1 | 8.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 2.0 | 6.0 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 2.0 | 10.0 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 2.0 | 13.7 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 1.8 | 1.8 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 1.8 | 9.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 1.8 | 14.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 1.7 | 5.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 1.7 | 6.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.7 | 13.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 1.7 | 5.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 1.7 | 18.7 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 1.7 | 13.4 | GO:0015705 | iodide transport(GO:0015705) |
| 1.6 | 6.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 1.6 | 4.9 | GO:0071529 | cementum mineralization(GO:0071529) |
| 1.6 | 8.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 1.6 | 4.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.6 | 6.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.6 | 4.8 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 1.6 | 4.8 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 1.6 | 4.7 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 1.5 | 16.7 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.5 | 12.0 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 1.4 | 13.0 | GO:0014744 | positive regulation of muscle adaptation(GO:0014744) |
| 1.4 | 11.5 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 1.4 | 10.0 | GO:0010269 | response to selenium ion(GO:0010269) |
| 1.4 | 18.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.3 | 8.0 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.3 | 7.8 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 1.3 | 22.9 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 1.2 | 6.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 1.2 | 3.7 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 1.2 | 4.9 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 1.2 | 6.0 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 1.1 | 4.5 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 1.1 | 4.5 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 1.1 | 18.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 1.1 | 3.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 1.1 | 3.2 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 1.1 | 3.2 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 1.1 | 5.3 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.1 | 6.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.0 | 7.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.0 | 7.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.0 | 8.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 1.0 | 17.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 1.0 | 5.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 1.0 | 7.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 1.0 | 8.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.0 | 14.1 | GO:0032196 | transposition(GO:0032196) |
| 1.0 | 1.9 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 1.0 | 4.8 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 1.0 | 1.9 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.9 | 16.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.9 | 7.6 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.9 | 4.7 | GO:0033504 | floor plate development(GO:0033504) |
| 0.9 | 8.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.9 | 10.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.9 | 12.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.9 | 5.3 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.9 | 4.3 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.9 | 3.4 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.9 | 12.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.8 | 12.7 | GO:0021513 | spinal cord dorsal/ventral patterning(GO:0021513) |
| 0.8 | 1.7 | GO:0042746 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.8 | 10.1 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.8 | 9.2 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.8 | 2.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.8 | 8.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.8 | 20.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.8 | 0.8 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.8 | 4.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.8 | 8.5 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.8 | 9.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.8 | 9.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.7 | 3.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.7 | 2.2 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.7 | 5.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.7 | 5.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.7 | 17.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.7 | 3.7 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.7 | 20.5 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.7 | 4.4 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.7 | 7.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) action potential propagation(GO:0098870) |
| 0.7 | 12.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.7 | 22.7 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.7 | 2.8 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.7 | 4.1 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.7 | 2.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.7 | 2.7 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.7 | 9.6 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.7 | 3.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.7 | 5.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.6 | 5.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.6 | 5.7 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.6 | 31.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.6 | 1.9 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.6 | 1.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.6 | 3.6 | GO:0002920 | regulation of humoral immune response(GO:0002920) regulation of complement activation(GO:0030449) regulation of protein activation cascade(GO:2000257) |
| 0.6 | 3.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.6 | 2.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.6 | 2.3 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.6 | 4.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.6 | 2.8 | GO:0015688 | response to mycotoxin(GO:0010046) iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.6 | 7.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 14.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.5 | 10.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.5 | 2.7 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.5 | 4.9 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.5 | 2.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.5 | 10.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 2.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.5 | 4.8 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.5 | 5.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.5 | 5.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 5.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.5 | 14.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.5 | 15.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.5 | 1.4 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.5 | 1.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.5 | 3.3 | GO:0046618 | drug export(GO:0046618) |
| 0.5 | 7.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.5 | 7.8 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.5 | 2.8 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.5 | 5.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.4 | 1.8 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.4 | 1.7 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.4 | 1.7 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.4 | 11.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.4 | 4.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.4 | 5.0 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.4 | 3.8 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.4 | 6.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.4 | 2.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.4 | 6.1 | GO:0045926 | negative regulation of cell growth(GO:0030308) negative regulation of growth(GO:0045926) |
| 0.4 | 4.5 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 2.4 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.4 | 1.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 2.4 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.4 | 10.3 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 1.6 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.4 | 4.6 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.4 | 0.8 | GO:1903059 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.4 | 4.2 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.4 | 1.1 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.4 | 4.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.4 | 58.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 7.2 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.4 | 3.5 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.3 | 3.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.3 | 1.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.3 | 4.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 0.9 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.3 | 8.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 6.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.3 | 11.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.3 | 8.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.3 | 0.9 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.3 | 1.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.3 | 3.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 11.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.3 | 2.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 11.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.3 | 5.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 1.6 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.3 | 2.2 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.3 | 1.1 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.3 | 33.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 7.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 7.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.2 | 12.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 18.1 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.2 | 2.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.2 | 2.8 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.2 | 2.8 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.2 | 7.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 4.6 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 8.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 0.6 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 | 6.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.2 | 7.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 | 11.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 1.0 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.2 | 3.9 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.2 | 7.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 2.8 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.2 | 1.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 6.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 6.0 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 1.4 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.2 | 10.1 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.2 | 4.5 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.2 | 3.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.2 | 3.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 4.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 1.7 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.2 | 6.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 23.2 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.2 | 2.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 1.8 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 4.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 3.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 2.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 5.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 5.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 2.3 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.2 | 5.1 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.1 | 3.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.1 | 2.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.5 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 6.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 2.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 8.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 5.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 4.0 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 1.7 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 5.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 3.9 | GO:0050776 | regulation of immune response(GO:0050776) |
| 0.1 | 2.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 4.4 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.1 | 1.3 | GO:0007350 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.1 | 2.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 5.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 1.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 10.1 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 7.5 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 5.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 6.3 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 2.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 2.2 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.4 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.1 | 5.5 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 1.4 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.8 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 2.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.8 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 5.1 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.5 | GO:0006701 | progesterone biosynthetic process(GO:0006701) Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 2.6 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.1 | 14.8 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.1 | 8.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.8 | GO:0045087 | innate immune response(GO:0045087) |
| 0.1 | 8.6 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.1 | 2.1 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 6.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 1.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 1.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 3.4 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.1 | 1.1 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 3.4 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 7.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 2.4 | GO:0007595 | lactation(GO:0007595) |
| 0.1 | 0.7 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 3.9 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 2.9 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 1.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 2.1 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 3.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 2.7 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.7 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 7.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.8 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 1.8 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.0 | 0.4 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 3.7 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 1.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.9 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 4.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 3.2 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 1.0 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 1.7 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 1.3 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0050890 | cognition(GO:0050890) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 15.3 | GO:0031417 | NatC complex(GO:0031417) |
| 3.3 | 9.8 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 2.3 | 11.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 2.2 | 11.0 | GO:0000801 | central element(GO:0000801) |
| 2.0 | 6.0 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 1.8 | 25.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.7 | 3.4 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 1.6 | 6.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 1.4 | 44.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 1.4 | 8.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.3 | 22.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 1.2 | 3.7 | GO:0098536 | deuterosome(GO:0098536) |
| 1.2 | 8.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 1.0 | 6.2 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.0 | 2.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 1.0 | 6.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.0 | 14.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 21.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.8 | 10.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.8 | 7.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.8 | 4.9 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.8 | 4.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.8 | 5.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.7 | 15.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.7 | 8.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.7 | 2.9 | GO:0005712 | chiasma(GO:0005712) |
| 0.7 | 7.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.7 | 13.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.7 | 16.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.7 | 7.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.6 | 9.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.6 | 2.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 14.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.6 | 12.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.6 | 10.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 6.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 2.9 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.5 | 6.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 7.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 9.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 2.9 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.5 | 3.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 2.9 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 1.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 22.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.5 | 12.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.5 | 9.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.4 | 11.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.4 | 3.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 2.8 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.4 | 7.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.4 | 5.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.4 | 20.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 10.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.4 | 2.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 0.7 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.3 | 4.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 34.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 48.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 1.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.3 | 11.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 4.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 3.0 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 10.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 22.1 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 10.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.3 | 22.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 3.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 22.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.3 | 31.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.3 | 6.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.3 | 11.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 11.9 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.3 | 1.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.3 | 7.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 7.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 9.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 2.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 3.6 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.2 | 2.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 1.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.2 | 10.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 10.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 11.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.2 | 49.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 4.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.7 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.2 | 5.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 12.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.2 | 10.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 14.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 4.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.2 | 1.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 0.9 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.2 | 1.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 4.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 20.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.4 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 28.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 3.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 14.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 4.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 12.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 4.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 7.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 9.0 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 13.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 4.4 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 4.8 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.1 | 1.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 26.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 184.7 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.1 | 106.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.3 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.9 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 30.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 40.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 3.8 | 11.5 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 3.8 | 41.7 | GO:0008430 | selenium binding(GO:0008430) |
| 3.5 | 20.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 3.2 | 13.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 2.7 | 10.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 2.7 | 8.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 2.6 | 10.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.4 | 7.3 | GO:0004040 | amidase activity(GO:0004040) fucose binding(GO:0042806) |
| 2.4 | 12.0 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 2.4 | 7.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 2.3 | 6.9 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 2.3 | 9.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 2.3 | 9.0 | GO:0004803 | transposase activity(GO:0004803) |
| 2.1 | 6.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 2.1 | 8.5 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 2.1 | 6.3 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 2.1 | 10.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.0 | 6.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.9 | 18.7 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 1.8 | 14.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.8 | 5.3 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 1.8 | 10.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 1.7 | 7.0 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 1.7 | 7.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.6 | 4.8 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 1.6 | 6.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 1.5 | 12.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.5 | 7.3 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.4 | 5.7 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 1.3 | 15.4 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 1.2 | 10.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.2 | 15.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 1.1 | 11.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.1 | 5.7 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 1.1 | 3.4 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 1.1 | 4.5 | GO:0016160 | amylase activity(GO:0016160) maltose alpha-glucosidase activity(GO:0032450) |
| 1.1 | 3.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 1.1 | 3.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 15.5 | GO:0031432 | titin binding(GO:0031432) |
| 1.1 | 6.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 1.1 | 3.2 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 1.0 | 5.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.0 | 10.1 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 1.0 | 9.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.0 | 3.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.0 | 6.9 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 1.0 | 17.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.9 | 2.8 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.9 | 15.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.9 | 2.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 8.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 23.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.9 | 5.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.8 | 15.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.8 | 5.9 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.8 | 2.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.8 | 20.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.8 | 4.9 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.8 | 7.9 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.8 | 4.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.8 | 3.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 3.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 4.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.7 | 5.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.7 | 1.5 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.7 | 4.4 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.7 | 2.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.7 | 10.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 4.9 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.7 | 14.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.7 | 4.8 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.7 | 10.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.7 | 14.0 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.6 | 31.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 4.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.6 | 1.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.6 | 3.7 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.6 | 5.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.6 | 7.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 7.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.6 | 4.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.6 | 9.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.6 | 10.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.6 | 16.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.6 | 40.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.6 | 21.6 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.6 | 1.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.5 | 1.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.5 | 1.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.5 | 8.6 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.5 | 3.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.5 | 2.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.5 | 9.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.5 | 5.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.5 | 7.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.5 | 1.4 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.5 | 2.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.4 | 4.5 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.4 | 20.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 8.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.4 | 4.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 1.7 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.4 | 5.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.4 | 3.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.4 | 1.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.4 | 4.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.4 | 4.7 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.4 | 8.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 8.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.4 | 2.3 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 5.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 6.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.4 | 2.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.4 | 4.6 | GO:0005372 | water transmembrane transporter activity(GO:0005372) water channel activity(GO:0015250) |
| 0.3 | 4.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 6.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 13.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 6.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 4.5 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.3 | 1.9 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.3 | 2.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.3 | 9.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 49.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 7.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 2.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 3.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 5.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 4.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.3 | 11.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.3 | 15.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.3 | 1.4 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.3 | 0.9 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.3 | 4.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 3.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.3 | 10.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 1.7 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 7.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.3 | 1.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.3 | 3.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 6.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 12.6 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.3 | 27.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 2.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.2 | 3.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 2.9 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 2.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 9.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 6.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 6.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 6.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 17.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.4 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 22.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 3.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.2 | 1.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 148.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.2 | 20.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 1.7 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.2 | 5.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 5.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 0.5 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 1.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 2.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.2 | 8.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.2 | 2.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 10.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 14.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 12.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 3.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 47.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 7.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 4.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 8.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.8 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 11.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 5.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 2.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 4.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 6.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 7.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 4.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 20.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 13.1 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.1 | 1.2 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 17.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 3.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 7.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 3.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 2.0 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.9 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 1.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.9 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 64.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.9 | 2.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 7.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.6 | 5.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.5 | 23.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.5 | 27.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 6.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 10.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.4 | 7.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.4 | 20.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 4.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 9.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 6.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 6.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 66.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 6.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.3 | 9.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 14.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.2 | 5.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 4.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 6.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 7.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 9.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 7.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 5.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 4.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 8.0 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 7.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 0.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 3.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 9.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.7 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 6.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 8.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 4.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 6.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 13.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.0 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 0.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 6.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 22.9 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 1.3 | 5.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.1 | 10.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.0 | 31.1 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 1.0 | 95.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 1.0 | 16.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.8 | 12.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.8 | 27.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 9.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.7 | 3.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.6 | 22.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.6 | 4.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 30.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.5 | 21.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.5 | 10.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 7.8 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.5 | 9.3 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.5 | 6.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.5 | 25.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.4 | 11.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 3.8 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.4 | 4.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 7.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 4.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 12.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 24.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 26.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.4 | 4.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 10.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 18.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 4.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 7.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 12.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 4.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.3 | 10.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 5.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 10.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 8.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 11.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 17.3 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.3 | 7.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 17.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 7.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.2 | 15.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 4.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 5.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 5.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.2 | 3.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 15.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 3.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.1 | 7.7 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 15.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 3.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 5.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 15.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 4.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 6.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 10.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 5.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 13.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 4.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.1 | REACTOME BIOLOGICAL OXIDATIONS | Genes involved in Biological oxidations |
| 0.1 | 2.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 4.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 8.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 5.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 6.9 | REACTOME GPCR DOWNSTREAM SIGNALING | Genes involved in GPCR downstream signaling |
| 0.1 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 2.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |