Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
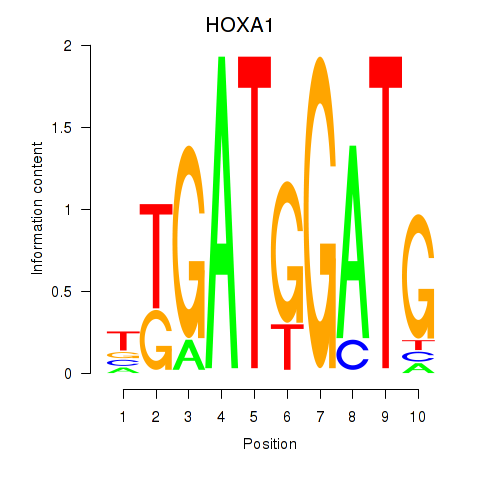
Results for HOXA1
Z-value: 0.67
Transcription factors associated with HOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA1
|
ENSG00000105991.7 | homeobox A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA1 | hg19_v2_chr7_-_27135591_27135658 | -0.04 | 5.2e-01 | Click! |
Activity profile of HOXA1 motif
Sorted Z-values of HOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_104152922 | 31.82 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr8_-_27468842 | 28.84 |
ENST00000523500.1
|
CLU
|
clusterin |
| chr5_-_42825983 | 28.79 |
ENST00000506577.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr8_-_27462822 | 23.03 |
ENST00000522098.1
|
CLU
|
clusterin |
| chr8_-_27468945 | 22.94 |
ENST00000405140.3
|
CLU
|
clusterin |
| chr8_-_27115931 | 19.27 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr20_-_23618582 | 18.46 |
ENST00000398411.1
ENST00000376925.3 |
CST3
|
cystatin C |
| chrX_-_13835147 | 18.39 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr8_-_27469196 | 18.10 |
ENST00000546343.1
ENST00000560566.1 |
CLU
|
clusterin |
| chr20_+_44036620 | 17.53 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_-_91539918 | 17.24 |
ENST00000548218.1
|
DCN
|
decorin |
| chr3_-_149688655 | 17.15 |
ENST00000461930.1
ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2
|
profilin 2 |
| chr8_-_27115903 | 16.65 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr2_+_149632783 | 16.54 |
ENST00000435030.1
|
KIF5C
|
kinesin family member 5C |
| chr6_+_121756809 | 15.98 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr11_-_5248294 | 15.61 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr11_-_72353451 | 15.37 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr20_+_44036900 | 14.43 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_+_166300084 | 14.14 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr9_-_101471479 | 13.56 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr2_+_24272576 | 13.42 |
ENST00000380986.4
ENST00000452109.1 |
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr19_+_5681011 | 13.25 |
ENST00000581893.1
ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr6_-_52705641 | 12.76 |
ENST00000370989.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr1_+_2005425 | 12.31 |
ENST00000461106.2
|
PRKCZ
|
protein kinase C, zeta |
| chr20_+_34742650 | 12.24 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr20_-_3154162 | 11.69 |
ENST00000360342.3
|
LZTS3
|
Homo sapiens leucine zipper, putative tumor suppressor family member 3 (LZTS3), mRNA. |
| chr4_+_113970772 | 11.57 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr16_+_15596123 | 10.95 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr11_-_104034827 | 10.51 |
ENST00000393158.2
|
PDGFD
|
platelet derived growth factor D |
| chr1_-_241520385 | 10.30 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr2_+_24272543 | 10.19 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr5_-_693500 | 10.16 |
ENST00000360578.5
|
TPPP
|
tubulin polymerization promoting protein |
| chr16_+_66914264 | 10.02 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr19_-_49944806 | 9.86 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr14_-_23822061 | 9.81 |
ENST00000397260.3
|
SLC22A17
|
solute carrier family 22, member 17 |
| chr8_+_22132847 | 9.31 |
ENST00000521356.1
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr11_-_104035088 | 9.26 |
ENST00000302251.5
|
PDGFD
|
platelet derived growth factor D |
| chr8_+_22132810 | 9.01 |
ENST00000356766.6
|
PIWIL2
|
piwi-like RNA-mediated gene silencing 2 |
| chr2_-_50574856 | 8.94 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr7_+_100136811 | 8.90 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr4_+_71587669 | 8.23 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_+_159557637 | 8.15 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr19_+_19144384 | 8.04 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr1_+_10271674 | 7.86 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr22_-_29711645 | 7.79 |
ENST00000401450.3
|
RASL10A
|
RAS-like, family 10, member A |
| chr2_-_44588694 | 7.78 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr19_+_13875316 | 7.54 |
ENST00000319545.8
ENST00000593245.1 ENST00000040663.6 |
MRI1
|
methylthioribose-1-phosphate isomerase 1 |
| chrX_-_57937067 | 7.41 |
ENST00000358697.4
|
ZXDA
|
zinc finger, X-linked, duplicated A |
| chr7_-_137028534 | 7.28 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr2_-_44588679 | 7.24 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr3_+_159570722 | 6.93 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr7_-_137028498 | 6.89 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr10_+_7745303 | 6.88 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr3_+_38537960 | 6.70 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr12_+_12878829 | 6.15 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr1_+_65730385 | 6.11 |
ENST00000263441.7
ENST00000395325.3 |
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr3_-_129612394 | 6.10 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr7_-_994302 | 6.08 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr5_+_175792459 | 6.01 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr9_-_97402413 | 5.58 |
ENST00000414122.1
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr1_-_13673511 | 5.57 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr17_-_42277203 | 5.52 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr6_+_151561085 | 5.49 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr16_+_75256507 | 5.48 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr10_+_7745232 | 5.32 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr19_-_39368887 | 5.28 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr17_-_3499125 | 5.25 |
ENST00000399759.3
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chrM_+_4431 | 5.15 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr19_-_40732594 | 5.10 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr9_+_10613163 | 5.06 |
ENST00000429581.2
|
RP11-87N24.2
|
RP11-87N24.2 |
| chr12_-_101604185 | 5.03 |
ENST00000536262.2
|
SLC5A8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr6_-_154677900 | 4.97 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr1_-_13452656 | 4.91 |
ENST00000376132.3
|
PRAMEF13
|
PRAME family member 13 |
| chr16_-_49890016 | 4.87 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chrX_+_57618269 | 4.85 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr10_-_79397391 | 4.84 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr16_+_29674540 | 4.75 |
ENST00000436527.1
ENST00000360121.3 ENST00000449759.1 |
SPN
QPRT
|
sialophorin quinolinate phosphoribosyltransferase |
| chr12_+_125478241 | 4.73 |
ENST00000341446.8
|
BRI3BP
|
BRI3 binding protein |
| chr1_-_40098672 | 4.65 |
ENST00000535435.1
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr12_-_121342170 | 4.58 |
ENST00000353487.2
|
SPPL3
|
signal peptide peptidase like 3 |
| chr12_-_11548496 | 4.54 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr22_+_29834572 | 4.51 |
ENST00000354373.2
|
RFPL1
|
ret finger protein-like 1 |
| chr3_+_4535155 | 4.50 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr10_+_60272814 | 4.40 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr1_+_205473720 | 4.40 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chrX_-_148586804 | 4.33 |
ENST00000428056.2
ENST00000340855.6 ENST00000370441.4 ENST00000370443.4 |
IDS
|
iduronate 2-sulfatase |
| chr6_+_7108210 | 4.32 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr8_-_91095099 | 4.31 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr20_+_8112824 | 4.28 |
ENST00000378641.3
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr4_-_90229142 | 4.25 |
ENST00000609438.1
|
GPRIN3
|
GPRIN family member 3 |
| chr17_-_3195876 | 4.18 |
ENST00000323404.1
|
OR3A1
|
olfactory receptor, family 3, subfamily A, member 1 |
| chr4_+_86396265 | 4.16 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_151561506 | 4.15 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr7_-_22233442 | 4.06 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr11_+_68228186 | 4.02 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_-_7913089 | 3.99 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr4_+_154178520 | 3.98 |
ENST00000433687.1
|
TRIM2
|
tripartite motif containing 2 |
| chr1_+_3614591 | 3.93 |
ENST00000378290.4
|
TP73
|
tumor protein p73 |
| chr11_+_61717336 | 3.76 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr11_+_61717279 | 3.73 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr1_+_162602244 | 3.70 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr5_-_177210399 | 3.68 |
ENST00000510276.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chrX_-_63005405 | 3.66 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr9_-_97402531 | 3.65 |
ENST00000415431.1
|
FBP1
|
fructose-1,6-bisphosphatase 1 |
| chr12_+_57610562 | 3.61 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr5_-_20575959 | 3.60 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr1_+_202976493 | 3.56 |
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A |
| chr1_-_12958101 | 3.55 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr1_-_45140074 | 3.52 |
ENST00000420706.1
ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53
|
transmembrane protein 53 |
| chr7_+_94139105 | 3.49 |
ENST00000297273.4
|
CASD1
|
CAS1 domain containing 1 |
| chr5_-_125930929 | 3.48 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr9_+_132962843 | 3.45 |
ENST00000458469.1
|
NCS1
|
neuronal calcium sensor 1 |
| chr5_+_36152179 | 3.38 |
ENST00000508514.1
ENST00000513151.1 ENST00000546211.1 |
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr12_-_89919965 | 3.33 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr7_+_79765071 | 3.33 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chrX_+_82763265 | 3.33 |
ENST00000373200.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr9_-_5304432 | 3.31 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr19_+_54641444 | 3.29 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr12_-_71533055 | 3.29 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr2_+_162016827 | 3.26 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr5_-_133702761 | 3.19 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr16_+_23847267 | 3.19 |
ENST00000321728.7
|
PRKCB
|
protein kinase C, beta |
| chr12_+_123459127 | 3.16 |
ENST00000397389.2
ENST00000538755.1 ENST00000536150.1 ENST00000545056.1 ENST00000545612.1 ENST00000538628.1 ENST00000545317.1 |
OGFOD2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr12_-_21927736 | 3.12 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chrX_+_15808569 | 3.04 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr10_-_105615164 | 3.03 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chrX_+_9217932 | 2.99 |
ENST00000432442.1
|
GS1-519E5.1
|
GS1-519E5.1 |
| chr1_+_12916941 | 2.98 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr11_+_2482661 | 2.98 |
ENST00000335475.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr17_-_44896047 | 2.95 |
ENST00000225512.5
|
WNT3
|
wingless-type MMTV integration site family, member 3 |
| chr13_-_20735178 | 2.95 |
ENST00000241125.3
|
GJA3
|
gap junction protein, alpha 3, 46kDa |
| chr19_-_42927251 | 2.88 |
ENST00000597001.1
|
LIPE
|
lipase, hormone-sensitive |
| chr17_-_15502111 | 2.86 |
ENST00000354433.3
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr12_-_96794330 | 2.80 |
ENST00000261211.3
|
CDK17
|
cyclin-dependent kinase 17 |
| chr13_+_92050928 | 2.80 |
ENST00000377067.3
|
GPC5
|
glypican 5 |
| chr11_+_63870660 | 2.78 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr11_-_2162162 | 2.77 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr15_-_58306295 | 2.72 |
ENST00000559517.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr5_+_159656437 | 2.67 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr3_+_4535025 | 2.66 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_-_51192661 | 2.47 |
ENST00000391813.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr14_+_65453432 | 2.45 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr16_+_71660079 | 2.38 |
ENST00000565261.1
ENST00000268485.3 ENST00000299952.4 |
MARVELD3
|
MARVEL domain containing 3 |
| chr19_-_33360647 | 2.38 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr9_-_89562104 | 2.32 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr17_+_26800296 | 2.32 |
ENST00000444914.3
ENST00000314669.5 |
SLC13A2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr12_-_96794143 | 2.31 |
ENST00000543119.2
|
CDK17
|
cyclin-dependent kinase 17 |
| chr12_+_54402790 | 2.31 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr7_-_44180884 | 2.31 |
ENST00000458240.1
ENST00000223364.3 |
MYL7
|
myosin, light chain 7, regulatory |
| chr12_+_8276495 | 2.31 |
ENST00000546339.1
|
CLEC4A
|
C-type lectin domain family 4, member A |
| chr9_+_87285257 | 2.28 |
ENST00000323115.4
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr1_+_17634689 | 2.20 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr6_+_25754927 | 2.19 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr2_-_79313973 | 2.18 |
ENST00000454188.1
|
REG1B
|
regenerating islet-derived 1 beta |
| chr7_-_126892303 | 2.12 |
ENST00000358373.3
|
GRM8
|
glutamate receptor, metabotropic 8 |
| chr5_-_141338627 | 2.11 |
ENST00000231484.3
|
PCDH12
|
protocadherin 12 |
| chr15_-_63674034 | 2.10 |
ENST00000344366.3
ENST00000422263.2 |
CA12
|
carbonic anhydrase XII |
| chr9_+_33795533 | 2.07 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr1_-_20306909 | 2.04 |
ENST00000375111.3
ENST00000400520.3 |
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chrX_+_37639302 | 2.04 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr10_-_134756030 | 2.03 |
ENST00000368586.5
ENST00000368582.2 |
TTC40
|
tetratricopeptide repeat domain 40 |
| chr11_+_60739249 | 1.96 |
ENST00000542157.1
ENST00000433107.2 ENST00000452451.2 ENST00000352009.5 |
CD6
|
CD6 molecule |
| chr8_+_143530791 | 1.94 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr2_+_65215604 | 1.90 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_125758865 | 1.90 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr17_+_61554413 | 1.80 |
ENST00000538928.1
ENST00000290866.4 ENST00000428043.1 |
ACE
|
angiotensin I converting enzyme |
| chr3_+_44916098 | 1.78 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr1_-_111061797 | 1.75 |
ENST00000369771.2
|
KCNA10
|
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr14_+_24779340 | 1.72 |
ENST00000533293.1
ENST00000543919.1 |
LTB4R2
|
leukotriene B4 receptor 2 |
| chr14_+_22948510 | 1.71 |
ENST00000390483.1
|
TRAJ56
|
T cell receptor alpha joining 56 |
| chr17_-_18950950 | 1.68 |
ENST00000284154.5
|
GRAP
|
GRB2-related adaptor protein |
| chr17_-_39041479 | 1.65 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr15_-_43513187 | 1.64 |
ENST00000540029.1
ENST00000441366.2 |
EPB42
|
erythrocyte membrane protein band 4.2 |
| chr8_-_40755333 | 1.63 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr20_+_52105495 | 1.61 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr19_+_54371114 | 1.61 |
ENST00000448420.1
ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM
|
myeloid-associated differentiation marker |
| chr2_+_8822113 | 1.60 |
ENST00000396290.1
ENST00000331129.3 |
ID2
|
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr6_-_47009996 | 1.58 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr13_+_102104952 | 1.57 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr16_-_55867146 | 1.56 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr17_+_72270429 | 1.54 |
ENST00000311014.6
|
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr6_+_42123141 | 1.54 |
ENST00000418175.1
ENST00000541991.1 ENST00000053469.4 ENST00000394237.1 ENST00000372963.1 |
GUCA1A
RP1-139D8.6
|
guanylate cyclase activator 1A (retina) RP1-139D8.6 |
| chr20_+_36012051 | 1.52 |
ENST00000373567.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr11_+_122753391 | 1.52 |
ENST00000307257.6
ENST00000227349.2 |
C11orf63
|
chromosome 11 open reading frame 63 |
| chr10_+_98741041 | 1.51 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr16_+_23847339 | 1.50 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr17_-_40264692 | 1.49 |
ENST00000591220.1
ENST00000251642.3 |
DHX58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr17_-_15501932 | 1.49 |
ENST00000583965.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr16_+_14805546 | 1.41 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr19_-_42192096 | 1.40 |
ENST00000602225.1
|
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr16_+_16472912 | 1.37 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr7_-_99332719 | 1.31 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr17_-_73401567 | 1.27 |
ENST00000392562.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr12_-_23737534 | 1.27 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr10_+_18689637 | 1.25 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr22_+_35695793 | 1.23 |
ENST00000456128.1
ENST00000449058.2 ENST00000411850.1 ENST00000425375.1 ENST00000436462.2 ENST00000382034.5 |
TOM1
|
target of myb1 (chicken) |
| chr19_+_58193337 | 1.23 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr19_-_15544099 | 1.22 |
ENST00000599910.2
|
WIZ
|
widely interspaced zinc finger motifs |
| chr7_+_97736197 | 1.22 |
ENST00000297293.5
|
LMTK2
|
lemur tyrosine kinase 2 |
| chr11_+_3666335 | 1.20 |
ENST00000250693.1
|
ART1
|
ADP-ribosyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.5 | 92.9 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 6.6 | 19.8 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 6.1 | 18.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 5.1 | 15.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 4.7 | 14.2 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 4.6 | 18.5 | GO:0051541 | elastin metabolic process(GO:0051541) negative regulation of blood vessel remodeling(GO:0060313) |
| 4.3 | 4.3 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 4.0 | 16.0 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 3.9 | 15.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 3.9 | 11.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 3.3 | 9.9 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 2.8 | 14.1 | GO:0030070 | insulin processing(GO:0030070) |
| 2.6 | 7.9 | GO:1904647 | response to rotenone(GO:1904647) |
| 2.4 | 23.6 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 2.3 | 9.2 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 2.2 | 10.9 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 2.1 | 12.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 2.0 | 9.8 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 1.8 | 18.4 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.7 | 5.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 1.6 | 4.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.6 | 17.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.6 | 4.7 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 1.4 | 4.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 1.4 | 17.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.4 | 7.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 1.2 | 4.7 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 1.1 | 8.9 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 4.0 | GO:0042320 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 1.0 | 2.9 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) Spemann organizer formation(GO:0060061) |
| 0.9 | 0.9 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.8 | 7.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.8 | 2.5 | GO:0050894 | determination of affect(GO:0050894) |
| 0.8 | 3.0 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.7 | 3.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.7 | 4.3 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.7 | 32.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.7 | 6.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.6 | 2.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.6 | 7.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.6 | 4.6 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.6 | 10.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.5 | 2.2 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.5 | 4.3 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.5 | 4.8 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.5 | 1.6 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.5 | 3.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.5 | 2.0 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.5 | 1.5 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.5 | 3.5 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.5 | 5.8 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.5 | 17.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.5 | 2.3 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.4 | 11.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.4 | 2.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 2.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 1.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.4 | 1.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.4 | 15.0 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.4 | 1.5 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.4 | 3.0 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.4 | 5.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 15.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.4 | 4.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.3 | 2.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.3 | 13.8 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.3 | 0.9 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.3 | 3.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.3 | 7.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.3 | 8.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.3 | 3.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.3 | 1.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.3 | 2.8 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 | 12.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 1.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.3 | 0.8 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.2 | 1.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 2.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.2 | 3.9 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.2 | 3.1 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.2 | 2.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.2 | 1.2 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 12.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 4.3 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.2 | 11.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 1.3 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.2 | 1.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.6 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 2.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.5 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 3.2 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.1 | 12.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.9 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 6.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 2.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 3.3 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 2.0 | GO:0001771 | immunological synapse formation(GO:0001771) positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.1 | 2.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 4.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.9 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 3.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 28.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 1.1 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 4.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 2.8 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.1 | 3.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 2.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 2.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 3.3 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 42.1 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 3.5 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 3.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 3.4 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 1.2 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 3.0 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 3.3 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.4 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 2.3 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 1.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.8 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 2.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 6.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 4.0 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.1 | GO:0061086 | regulation of mRNA export from nucleus(GO:0010793) negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.3 | GO:0008212 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.0 | 0.5 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 1.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.8 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 92.9 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 4.5 | 13.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 3.9 | 15.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.7 | 18.3 | GO:0071546 | perinucleolar chromocenter(GO:0010370) pi-body(GO:0071546) |
| 1.5 | 12.3 | GO:0045179 | apical cortex(GO:0045179) |
| 1.3 | 16.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.2 | 9.9 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 1.2 | 17.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.1 | 16.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 1.0 | 3.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 1.0 | 10.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.8 | 7.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.7 | 4.6 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.6 | 2.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.6 | 3.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.5 | 24.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.4 | 32.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.4 | 2.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 3.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 22.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 3.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 37.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.3 | 26.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 2.9 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.3 | 5.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 10.9 | GO:0043034 | costamere(GO:0043034) |
| 0.3 | 7.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 53.5 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 37.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 1.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 7.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 2.9 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 2.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 6.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 9.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 7.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 10.7 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 3.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 2.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 9.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 15.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 12.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 3.1 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 13.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 4.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 8.0 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 4.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 5.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 4.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 1.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.3 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 18.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 4.7 | 23.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 4.6 | 92.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 4.5 | 13.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 3.5 | 14.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.7 | 16.0 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 2.6 | 15.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 2.6 | 15.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 2.5 | 9.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.3 | 9.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 1.8 | 7.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.7 | 10.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.6 | 4.7 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 1.6 | 4.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.3 | 5.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 1.3 | 29.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 1.2 | 4.7 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.1 | 4.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 1.0 | 3.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 1.0 | 6.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 1.0 | 2.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.9 | 10.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.9 | 19.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.8 | 2.5 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.8 | 2.3 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.7 | 3.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.7 | 4.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.7 | 4.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.7 | 7.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 13.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.6 | 2.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.6 | 3.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.6 | 9.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.6 | 18.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.6 | 8.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 3.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.5 | 3.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.5 | 7.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.5 | 2.3 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.5 | 5.0 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.4 | 2.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 3.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 1.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.4 | 7.9 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.4 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 17.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 6.7 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.4 | 2.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 1.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.3 | 3.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.3 | 1.6 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.3 | 4.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.3 | 1.5 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 0.9 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.3 | 2.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 3.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 2.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.3 | 3.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 2.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 1.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.3 | 6.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 12.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 7.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 1.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 3.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 0.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 18.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 15.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.8 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.2 | 4.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 0.8 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 2.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 3.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 2.7 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.2 | 4.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 1.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 2.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 0.9 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 2.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.8 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.9 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 12.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 3.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 2.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 6.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 5.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 2.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 5.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 2.8 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 0.9 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 28.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 4.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 1.3 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 18.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 3.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 8.3 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 2.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 1.0 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 3.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 4.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.2 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 87.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.5 | 33.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 15.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 18.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 4.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 17.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.3 | 13.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 5.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 16.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 11.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.2 | 14.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.2 | 4.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 6.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 34.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 3.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 3.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 4.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 16.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.9 | 15.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.8 | 21.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.6 | 10.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.6 | 90.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.5 | 14.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 12.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 9.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 4.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 15.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 17.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 12.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 7.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 10.4 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 3.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 3.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 7.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 2.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 3.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 6.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 9.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 4.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.2 | 2.8 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 6.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 2.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 2.8 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 6.0 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 4.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 4.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 2.3 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 3.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 6.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.4 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 1.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 4.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 8.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |