Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
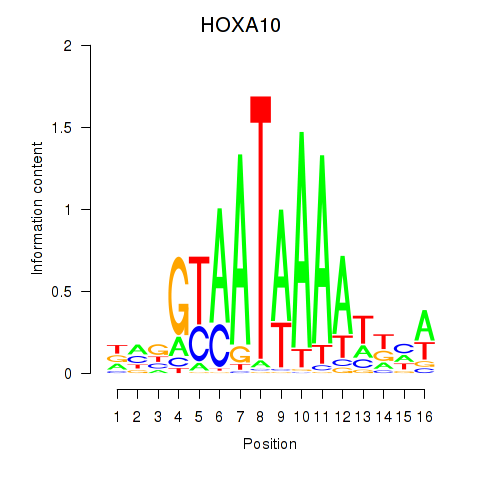
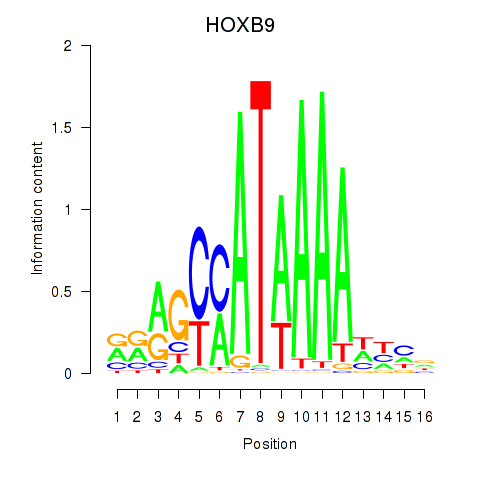
Results for HOXA10_HOXB9
Z-value: 0.07
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA10
|
ENSG00000253293.3 | homeobox A10 |
|
HOXB9
|
ENSG00000170689.8 | homeobox B9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27213893_27213954 | -0.21 | 1.6e-03 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | -0.13 | 6.5e-02 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_84630645 | 8.47 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_-_55541227 | 8.33 |
ENST00000566877.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr10_-_73848086 | 7.61 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr10_-_73848531 | 7.11 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr3_-_18480260 | 6.61 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr8_-_81083890 | 6.15 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chr4_-_71532339 | 5.87 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr2_-_51259292 | 5.80 |
ENST00000401669.2
|
NRXN1
|
neurexin 1 |
| chr4_+_71248795 | 5.74 |
ENST00000304915.3
|
SMR3B
|
submaxillary gland androgen regulated protein 3B |
| chr2_-_51259229 | 5.74 |
ENST00000405472.3
|
NRXN1
|
neurexin 1 |
| chr9_+_2029019 | 5.55 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_-_60097524 | 5.30 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr14_-_60097297 | 5.26 |
ENST00000395090.1
|
RTN1
|
reticulon 1 |
| chr12_+_54892550 | 5.08 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr1_-_160231451 | 4.82 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr4_-_57522598 | 4.61 |
ENST00000553379.2
|
HOPX
|
HOP homeobox |
| chr6_+_158733692 | 4.61 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr11_-_104972158 | 4.43 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr4_-_57522673 | 4.31 |
ENST00000381255.3
ENST00000317745.7 ENST00000555760.2 ENST00000556614.2 |
HOPX
|
HOP homeobox |
| chr12_+_25205568 | 3.95 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr18_-_5396271 | 3.93 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr4_+_70894130 | 3.89 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr10_+_94451574 | 3.83 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr6_+_32812568 | 3.82 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_-_90049878 | 3.69 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_+_67586465 | 3.67 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr12_-_90049828 | 3.63 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr4_-_57547870 | 3.62 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr4_+_70861647 | 3.59 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr6_+_6588316 | 3.46 |
ENST00000379953.2
|
LY86
|
lymphocyte antigen 86 |
| chr1_+_12524965 | 3.38 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr6_+_12958137 | 3.33 |
ENST00000457702.2
ENST00000379345.2 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr4_-_100356551 | 3.18 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_-_104905840 | 3.15 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr9_+_137979506 | 3.10 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr4_+_70916119 | 3.03 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr4_+_68424434 | 2.96 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr12_+_25205666 | 2.89 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr5_-_88119580 | 2.86 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr12_+_25205446 | 2.85 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr17_-_10452929 | 2.78 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr22_-_23922410 | 2.48 |
ENST00000249053.3
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr12_-_118628350 | 2.46 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr5_-_58295712 | 2.30 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_90198535 | 2.19 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr5_-_130500922 | 2.18 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr17_-_29624343 | 2.18 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr11_-_102576537 | 2.13 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr9_+_134065506 | 2.09 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr16_-_15180257 | 2.04 |
ENST00000540462.1
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr1_-_152386732 | 2.00 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr11_-_3400442 | 1.97 |
ENST00000429541.2
ENST00000532539.1 |
ZNF195
|
zinc finger protein 195 |
| chr17_+_48823975 | 1.97 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr11_-_3400330 | 1.91 |
ENST00000427810.2
ENST00000005082.9 ENST00000534569.1 ENST00000438262.2 ENST00000528796.1 ENST00000528410.1 ENST00000529678.1 ENST00000354599.6 ENST00000526601.1 ENST00000525502.1 ENST00000533036.1 ENST00000399602.4 |
ZNF195
|
zinc finger protein 195 |
| chr10_-_103599591 | 1.81 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr1_-_226926864 | 1.77 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr3_-_50375657 | 1.75 |
ENST00000395126.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr8_-_81083731 | 1.72 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr3_-_58196688 | 1.70 |
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr9_-_95087838 | 1.68 |
ENST00000442668.2
ENST00000421075.2 ENST00000536624.1 |
NOL8
|
nucleolar protein 8 |
| chr19_-_29704448 | 1.67 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr21_+_34398153 | 1.62 |
ENST00000382357.3
ENST00000430860.1 ENST00000333337.3 |
OLIG2
|
oligodendrocyte lineage transcription factor 2 |
| chr5_+_43121698 | 1.59 |
ENST00000505606.2
ENST00000509634.1 ENST00000509341.1 |
ZNF131
|
zinc finger protein 131 |
| chr18_-_53069419 | 1.58 |
ENST00000570177.2
|
TCF4
|
transcription factor 4 |
| chr11_+_10471836 | 1.57 |
ENST00000444303.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr8_+_1993152 | 1.57 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr1_+_73771844 | 1.56 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr8_+_95565947 | 1.55 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr5_+_140261703 | 1.54 |
ENST00000409494.1
ENST00000289272.2 |
PCDHA13
|
protocadherin alpha 13 |
| chr12_-_54071181 | 1.54 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr8_+_1993173 | 1.53 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr6_+_83903061 | 1.52 |
ENST00000369724.4
ENST00000539997.1 |
RWDD2A
|
RWD domain containing 2A |
| chr9_+_91933726 | 1.51 |
ENST00000534113.2
|
SECISBP2
|
SECIS binding protein 2 |
| chrX_-_122756660 | 1.51 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr1_+_154401791 | 1.49 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr20_-_8000426 | 1.47 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr4_-_140222358 | 1.38 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr14_-_25479811 | 1.35 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr1_+_153003671 | 1.34 |
ENST00000307098.4
|
SPRR1B
|
small proline-rich protein 1B |
| chr7_+_24323782 | 1.34 |
ENST00000242152.2
ENST00000407573.1 |
NPY
|
neuropeptide Y |
| chr4_+_71337834 | 1.33 |
ENST00000304887.5
|
MUC7
|
mucin 7, secreted |
| chr10_+_96698406 | 1.33 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr8_-_95220775 | 1.32 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr13_-_79979919 | 1.30 |
ENST00000267229.7
|
RBM26
|
RNA binding motif protein 26 |
| chr2_+_208423840 | 1.29 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr4_+_71226468 | 1.29 |
ENST00000226460.4
|
SMR3A
|
submaxillary gland androgen regulated protein 3A |
| chr6_+_42584847 | 1.28 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr15_-_22448819 | 1.26 |
ENST00000604066.1
|
IGHV1OR15-1
|
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr12_-_81992111 | 1.25 |
ENST00000443686.3
ENST00000407050.4 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr13_-_79979952 | 1.25 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr1_+_229440129 | 1.25 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr2_-_40657397 | 1.24 |
ENST00000408028.2
ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr2_+_90192768 | 1.23 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr4_-_90759440 | 1.23 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_-_135962876 | 1.22 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chr13_-_47012325 | 1.21 |
ENST00000409879.2
|
KIAA0226L
|
KIAA0226-like |
| chr18_-_52989217 | 1.18 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr2_+_173686303 | 1.16 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr5_+_140165876 | 1.16 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr20_-_7921090 | 1.14 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chrX_+_105937068 | 1.13 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr18_-_52989525 | 1.10 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr14_+_53173910 | 1.09 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr9_-_179018 | 1.07 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr14_+_53173890 | 1.07 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr15_-_58571445 | 1.06 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr16_-_21663919 | 1.06 |
ENST00000569602.1
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr10_+_133747955 | 1.04 |
ENST00000455566.1
|
PPP2R2D
|
protein phosphatase 2, regulatory subunit B, delta |
| chr9_+_125703282 | 1.02 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr22_-_23922448 | 0.99 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr3_+_169629354 | 0.99 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr4_+_113568207 | 0.98 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr7_-_142232071 | 0.97 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr2_-_176046391 | 0.97 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr3_+_157828152 | 0.93 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr4_-_87281224 | 0.92 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_57146095 | 0.91 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr8_+_128426535 | 0.90 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr9_+_70856397 | 0.89 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr8_+_132952112 | 0.86 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr1_+_74701062 | 0.86 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr6_+_26156551 | 0.85 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr5_-_176433693 | 0.85 |
ENST00000507513.1
ENST00000511320.1 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr1_+_22379179 | 0.84 |
ENST00000315554.8
ENST00000421089.2 |
CDC42
|
cell division cycle 42 |
| chr20_-_34287103 | 0.84 |
ENST00000374085.1
ENST00000419569.1 |
NFS1
|
NFS1 cysteine desulfurase |
| chr11_-_118972575 | 0.83 |
ENST00000432443.2
|
DPAGT1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr19_-_3500635 | 0.83 |
ENST00000250937.3
|
DOHH
|
deoxyhypusine hydroxylase/monooxygenase |
| chr19_+_19626531 | 0.83 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr4_-_76957214 | 0.82 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chrX_-_110655306 | 0.82 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr1_-_39339777 | 0.82 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr5_-_20575959 | 0.81 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr19_+_21265028 | 0.81 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr10_+_88428206 | 0.81 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr3_-_123710199 | 0.79 |
ENST00000184183.4
|
ROPN1
|
rhophilin associated tail protein 1 |
| chr5_+_135496675 | 0.78 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr9_+_42671887 | 0.77 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr3_-_172241250 | 0.77 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr2_-_152382500 | 0.74 |
ENST00000434685.1
|
NEB
|
nebulin |
| chr13_-_103719196 | 0.72 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr11_-_13517565 | 0.72 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr8_-_8318847 | 0.72 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr14_-_106453155 | 0.72 |
ENST00000390594.2
|
IGHV1-2
|
immunoglobulin heavy variable 1-2 |
| chr6_-_137539651 | 0.72 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr7_+_57509877 | 0.71 |
ENST00000420713.1
|
ZNF716
|
zinc finger protein 716 |
| chr4_+_71263599 | 0.69 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr1_-_7913089 | 0.69 |
ENST00000361696.5
|
UTS2
|
urotensin 2 |
| chr17_-_10560619 | 0.69 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr5_+_140514782 | 0.67 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr17_-_46806540 | 0.67 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr9_-_69262509 | 0.63 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr1_-_54355430 | 0.63 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr22_-_22292934 | 0.62 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chrX_-_15619076 | 0.61 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr16_-_21663950 | 0.60 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr12_-_71551868 | 0.59 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr11_-_104827425 | 0.58 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_31456874 | 0.57 |
ENST00000541626.1
|
EHD3
|
EH-domain containing 3 |
| chr12_-_8765446 | 0.57 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr16_+_25123041 | 0.55 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr11_+_61717336 | 0.54 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr19_+_1440838 | 0.52 |
ENST00000594262.1
|
AC027307.3
|
Uncharacterized protein |
| chr15_+_75639296 | 0.52 |
ENST00000564500.1
ENST00000355059.4 ENST00000566752.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr18_+_19192228 | 0.52 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr14_+_79745746 | 0.52 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr13_-_46626847 | 0.51 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr20_-_50384864 | 0.50 |
ENST00000311637.5
ENST00000402822.1 |
ATP9A
|
ATPase, class II, type 9A |
| chr19_+_12175504 | 0.49 |
ENST00000439326.3
|
ZNF844
|
zinc finger protein 844 |
| chr2_-_154335300 | 0.49 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chrX_-_73072534 | 0.49 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr2_+_208423891 | 0.48 |
ENST00000448277.1
ENST00000457101.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_+_67835517 | 0.46 |
ENST00000395476.2
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr11_+_61717279 | 0.46 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr6_-_109804412 | 0.45 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr18_+_42260861 | 0.45 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr12_+_57810198 | 0.44 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr2_+_27440229 | 0.43 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr9_+_82187630 | 0.42 |
ENST00000265284.6
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr22_+_24407642 | 0.42 |
ENST00000454754.1
ENST00000263119.5 |
CABIN1
|
calcineurin binding protein 1 |
| chr2_-_209010874 | 0.42 |
ENST00000260988.4
|
CRYGB
|
crystallin, gamma B |
| chr6_-_8102714 | 0.42 |
ENST00000502429.1
ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr1_+_67773044 | 0.41 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr10_+_91152303 | 0.41 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr2_+_161993412 | 0.41 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr9_+_82187487 | 0.40 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr8_+_76452097 | 0.38 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr1_-_143767881 | 0.37 |
ENST00000419275.1
|
PPIAL4G
|
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr14_-_51027838 | 0.36 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_220263096 | 0.36 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr17_-_39553844 | 0.35 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr9_-_104145795 | 0.34 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr1_+_151739131 | 0.34 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr1_+_241695670 | 0.33 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr9_+_103947311 | 0.33 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr1_+_117963209 | 0.32 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr11_+_120973375 | 0.32 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr2_+_234668894 | 0.31 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr22_-_22337146 | 0.28 |
ENST00000398793.2
|
TOP3B
|
topoisomerase (DNA) III beta |
| chr2_+_220144052 | 0.28 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr19_+_48497962 | 0.28 |
ENST00000596043.1
ENST00000597519.1 |
ELSPBP1
|
epididymal sperm binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 1.7 | 8.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.5 | 7.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.4 | 11.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.4 | 8.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 1.3 | 3.8 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 1.3 | 5.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 1.1 | 3.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 1.0 | 3.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.7 | 2.9 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.7 | 14.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.7 | 1.3 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.6 | 3.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.6 | 6.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 | 12.5 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.5 | 1.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.5 | 1.5 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.4 | 3.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 3.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.3 | 3.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.3 | 0.9 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.8 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.3 | 2.0 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.3 | 0.8 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.3 | 2.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 | 1.6 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.3 | 1.8 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 1.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.3 | 2.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.3 | 1.8 | GO:0033029 | neutrophil apoptotic process(GO:0001781) regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.2 | 1.2 | GO:1903282 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.7 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 2.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 3.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.2 | 0.8 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 1.7 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.2 | 0.6 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.2 | 1.3 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.2 | 1.3 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.2 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.7 | GO:0042320 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) |
| 0.2 | 6.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 1.5 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 0.7 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 1.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.2 | 1.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 1.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.4 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 1.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.8 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.4 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.5 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 1.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.5 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 2.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.7 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.8 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.4 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 4.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 2.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.6 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.3 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.1 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.8 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 2.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.9 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 1.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 9.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 4.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.6 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.6 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.3 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.2 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 1.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.6 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 2.1 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 4.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 2.2 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.3 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 5.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 3.8 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 1.3 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.9 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 1.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 2.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:2000774 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 3.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 2.1 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.8 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 2.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.8 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.7 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.3 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 2.0 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.5 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 2.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.7 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.9 | 2.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.9 | 3.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.7 | 2.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.7 | 8.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 3.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.3 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.4 | 8.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 1.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 5.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 7.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 1.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 1.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 5.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 1.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.6 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 2.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 7.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 1.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 3.8 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 0.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 3.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 4.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 2.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 9.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 11.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 4.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.2 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.1 | 0.8 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.5 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.1 | 11.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 7.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 8.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 3.3 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.9 | 4.3 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.8 | 12.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.7 | 2.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.6 | 8.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.5 | 13.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 8.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 1.5 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.5 | 3.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.5 | 3.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.4 | 1.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.3 | 8.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 1.3 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.3 | 8.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 3.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 1.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 0.8 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 0.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 1.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.3 | 1.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 7.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 1.5 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 1.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 1.8 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.8 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.2 | 2.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 2.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 1.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.2 | 1.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.4 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 0.7 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 3.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 3.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.5 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 2.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 5.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 4.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.1 | 0.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 2.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.8 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 4.8 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 2.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.4 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 7.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 4.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 1.0 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 3.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 3.5 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 3.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 14.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 12.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 1.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 12.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 6.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 4.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 3.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 8.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 10.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 3.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 7.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 7.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 6.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 6.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 2.0 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 3.5 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |