Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
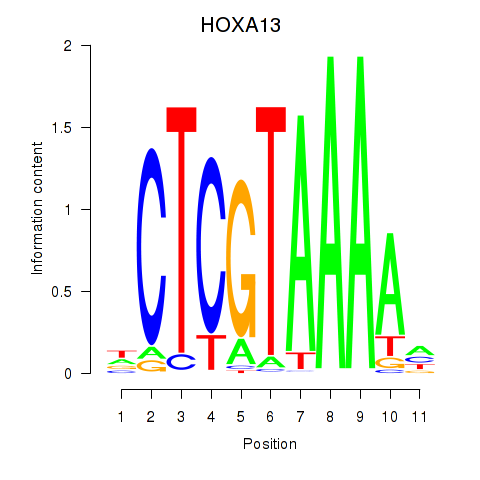
Results for HOXA13
Z-value: 0.32
Transcription factors associated with HOXA13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA13
|
ENSG00000106031.6 | homeobox A13 |
Activity profile of HOXA13 motif
Sorted Z-values of HOXA13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 13.82 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr12_-_91539918 | 12.96 |
ENST00000548218.1
|
DCN
|
decorin |
| chr6_+_121756809 | 11.91 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr3_-_123339418 | 10.06 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_123339343 | 9.60 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr12_-_91546926 | 8.10 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_-_112127981 | 6.93 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr5_-_79287060 | 6.81 |
ENST00000512560.1
ENST00000509852.1 ENST00000512528.1 |
MTX3
|
metaxin 3 |
| chr12_+_6309517 | 6.46 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr1_+_43766642 | 6.07 |
ENST00000372476.3
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr12_+_120105558 | 5.67 |
ENST00000229328.5
ENST00000541640.1 |
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr8_+_133879193 | 5.58 |
ENST00000377869.1
ENST00000220616.4 |
TG
|
thyroglobulin |
| chr6_+_151042224 | 5.53 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr12_-_12491608 | 5.46 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr4_+_106631966 | 5.16 |
ENST00000360505.5
ENST00000510865.1 ENST00000509336.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr19_+_16999654 | 4.63 |
ENST00000248076.3
|
F2RL3
|
coagulation factor II (thrombin) receptor-like 3 |
| chr6_+_19837592 | 4.48 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr8_-_86290333 | 4.33 |
ENST00000521846.1
ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1
|
carbonic anhydrase I |
| chr12_+_12878829 | 3.99 |
ENST00000326765.6
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr12_-_71182695 | 3.95 |
ENST00000342084.4
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr9_+_137979506 | 3.88 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr14_-_92414055 | 3.83 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr18_+_39766626 | 3.81 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr8_-_95274536 | 3.80 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr6_+_131894284 | 3.74 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr11_+_61891445 | 3.64 |
ENST00000394818.3
ENST00000533896.1 ENST00000278849.4 |
INCENP
|
inner centromere protein antigens 135/155kDa |
| chr3_+_101546827 | 3.56 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr9_+_706842 | 3.50 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr11_-_114466477 | 3.41 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr2_-_177502254 | 3.21 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chrX_+_35816459 | 3.19 |
ENST00000399988.1
ENST00000399992.1 ENST00000399987.1 ENST00000399989.1 |
MAGEB16
|
melanoma antigen family B, 16 |
| chr2_-_32390801 | 3.04 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr6_-_11807277 | 3.02 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr16_+_66914264 | 3.00 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr3_-_99569821 | 2.84 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr17_-_40021656 | 2.83 |
ENST00000319121.3
|
KLHL11
|
kelch-like family member 11 |
| chr1_-_25756638 | 2.82 |
ENST00000349320.3
|
RHCE
|
Rh blood group, CcEe antigens |
| chr4_+_70894130 | 2.75 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr12_-_53074182 | 2.70 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr20_-_52687059 | 2.69 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr3_-_114477787 | 2.66 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_+_95401877 | 2.58 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr4_+_74269956 | 2.57 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr11_-_114466471 | 2.57 |
ENST00000424261.2
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr7_+_95401851 | 2.53 |
ENST00000447467.2
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr5_-_138210977 | 2.50 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr12_+_26274917 | 2.49 |
ENST00000538142.1
|
SSPN
|
sarcospan |
| chr14_+_29236269 | 2.41 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr15_-_99789736 | 2.37 |
ENST00000560235.1
ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr6_+_148663729 | 2.28 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_+_13910194 | 2.25 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr12_-_8693539 | 2.24 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr4_-_89978299 | 2.18 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr5_-_19988339 | 2.11 |
ENST00000382275.1
|
CDH18
|
cadherin 18, type 2 |
| chr3_-_160823158 | 2.04 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr11_-_104480019 | 2.03 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr7_+_80275752 | 2.01 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr8_-_9008206 | 2.00 |
ENST00000310455.3
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_-_149093499 | 1.99 |
ENST00000472441.1
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr19_+_8483272 | 1.95 |
ENST00000602117.1
|
MARCH2
|
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr19_-_44809121 | 1.95 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr5_+_66254698 | 1.94 |
ENST00000405643.1
ENST00000407621.1 ENST00000432426.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_91933726 | 1.93 |
ENST00000534113.2
|
SECISBP2
|
SECIS binding protein 2 |
| chr11_+_94706804 | 1.92 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr3_-_126373929 | 1.91 |
ENST00000523403.1
ENST00000524230.2 |
TXNRD3
|
thioredoxin reductase 3 |
| chr7_+_80275621 | 1.91 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr10_-_50970322 | 1.89 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chrX_-_38186775 | 1.88 |
ENST00000339363.3
ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR
|
retinitis pigmentosa GTPase regulator |
| chr19_+_16830774 | 1.83 |
ENST00000524140.2
|
NWD1
|
NACHT and WD repeat domain containing 1 |
| chr4_-_17513702 | 1.81 |
ENST00000428702.2
ENST00000508623.1 ENST00000513615.1 |
QDPR
|
quinoid dihydropteridine reductase |
| chr3_-_160822858 | 1.79 |
ENST00000488170.1
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr14_-_71609965 | 1.78 |
ENST00000602957.1
|
RP6-91H8.3
|
RP6-91H8.3 |
| chr3_-_160823040 | 1.78 |
ENST00000484127.1
ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr16_+_82090028 | 1.78 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr1_-_31845914 | 1.76 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr18_-_5544241 | 1.76 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr16_-_21289627 | 1.76 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr13_-_36429763 | 1.75 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr11_+_101785727 | 1.74 |
ENST00000263468.8
|
KIAA1377
|
KIAA1377 |
| chr18_-_52989217 | 1.73 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr11_-_124670550 | 1.72 |
ENST00000239614.4
|
MSANTD2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr1_+_173793777 | 1.71 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr1_-_216978709 | 1.69 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr17_+_16284104 | 1.69 |
ENST00000577958.1
ENST00000302182.3 ENST00000577640.1 |
UBB
|
ubiquitin B |
| chr22_+_21400229 | 1.65 |
ENST00000342608.4
ENST00000543388.1 ENST00000442047.1 |
AC002472.13
|
Leucine-rich repeat-containing protein LOC400891 |
| chr2_-_241737128 | 1.63 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr4_+_70861647 | 1.61 |
ENST00000246895.4
ENST00000381060.2 |
STATH
|
statherin |
| chr22_+_39916558 | 1.56 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr19_-_18632861 | 1.54 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr14_+_100485712 | 1.54 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr11_-_85565906 | 1.53 |
ENST00000544076.1
|
AP000974.1
|
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
| chr19_-_47987419 | 1.52 |
ENST00000536339.1
ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN
|
kaptin (actin binding protein) |
| chr22_+_18632666 | 1.52 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr19_+_35773242 | 1.50 |
ENST00000222304.3
|
HAMP
|
hepcidin antimicrobial peptide |
| chr3_+_190333097 | 1.50 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr4_+_95679072 | 1.47 |
ENST00000515059.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr4_+_72052964 | 1.46 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr4_-_155533787 | 1.44 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr19_+_859654 | 1.43 |
ENST00000592860.1
|
CFD
|
complement factor D (adipsin) |
| chrX_+_37850026 | 1.43 |
ENST00000341016.3
|
CXorf27
|
chromosome X open reading frame 27 |
| chr2_+_33359646 | 1.42 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr7_-_27169801 | 1.38 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr7_+_80275953 | 1.38 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_+_33359687 | 1.36 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr22_+_38142235 | 1.34 |
ENST00000407319.2
ENST00000403663.2 ENST00000428075.1 |
TRIOBP
|
TRIO and F-actin binding protein |
| chr1_+_202431859 | 1.33 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr16_+_75252878 | 1.33 |
ENST00000361017.4
|
CTRB1
|
chymotrypsinogen B1 |
| chr11_+_7110165 | 1.29 |
ENST00000306904.5
|
RBMXL2
|
RNA binding motif protein, X-linked-like 2 |
| chr21_+_39644172 | 1.29 |
ENST00000398932.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr17_+_16284399 | 1.29 |
ENST00000535788.1
|
UBB
|
ubiquitin B |
| chr11_+_61015594 | 1.28 |
ENST00000451616.2
|
PGA5
|
pepsinogen 5, group I (pepsinogen A) |
| chr9_-_21305312 | 1.27 |
ENST00000259555.4
|
IFNA5
|
interferon, alpha 5 |
| chr16_-_66764119 | 1.26 |
ENST00000569320.1
|
DYNC1LI2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr4_-_17513851 | 1.25 |
ENST00000281243.5
|
QDPR
|
quinoid dihydropteridine reductase |
| chr19_+_54369608 | 1.21 |
ENST00000336967.3
|
MYADM
|
myeloid-associated differentiation marker |
| chr21_+_39644305 | 1.20 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_10590238 | 1.20 |
ENST00000256178.3
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr11_+_62037622 | 1.20 |
ENST00000227918.2
ENST00000525380.1 |
SCGB2A2
|
secretoglobin, family 2A, member 2 |
| chr5_+_140535577 | 1.20 |
ENST00000539533.1
|
PCDHB17
|
Protocadherin-psi1; Uncharacterized protein |
| chr21_+_39644395 | 1.20 |
ENST00000398934.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_+_129159039 | 1.19 |
ENST00000507564.1
ENST00000431818.2 ENST00000504021.1 ENST00000349441.2 ENST00000348417.2 ENST00000440957.2 |
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr21_+_39628655 | 1.16 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_-_78808617 | 1.15 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr7_-_27224795 | 1.14 |
ENST00000006015.3
|
HOXA11
|
homeobox A11 |
| chr1_-_46642154 | 1.13 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr2_+_182756615 | 1.13 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr2_+_33661382 | 1.11 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_+_78769549 | 1.10 |
ENST00000370758.1
|
PTGFR
|
prostaglandin F receptor (FP) |
| chr2_-_86850949 | 1.09 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr16_-_90096309 | 1.09 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr6_+_73076432 | 1.08 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr4_+_25657444 | 1.05 |
ENST00000504570.1
ENST00000382051.3 |
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr15_-_88799661 | 1.05 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr1_+_78511586 | 1.05 |
ENST00000370759.3
|
GIPC2
|
GIPC PDZ domain containing family, member 2 |
| chr18_-_52989525 | 1.04 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr9_-_98269699 | 1.04 |
ENST00000429896.2
|
PTCH1
|
patched 1 |
| chr20_+_55204351 | 1.04 |
ENST00000201031.2
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr1_-_207224307 | 1.01 |
ENST00000315927.4
|
YOD1
|
YOD1 deubiquitinase |
| chr1_-_16763911 | 0.99 |
ENST00000375577.1
ENST00000335496.1 |
SPATA21
|
spermatogenesis associated 21 |
| chr19_+_13858593 | 0.99 |
ENST00000221554.8
|
CCDC130
|
coiled-coil domain containing 130 |
| chr17_-_39553844 | 0.99 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr4_+_70796784 | 0.97 |
ENST00000246891.4
ENST00000444405.3 |
CSN1S1
|
casein alpha s1 |
| chr14_+_50234309 | 0.96 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr1_-_28177255 | 0.93 |
ENST00000601459.1
|
AL109927.1
|
HCG2032222; PRO2047; Uncharacterized protein |
| chr4_+_90823130 | 0.93 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr21_+_39644214 | 0.93 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr14_-_102706934 | 0.92 |
ENST00000523231.1
ENST00000524370.1 ENST00000517966.1 |
MOK
|
MOK protein kinase |
| chr5_+_67584174 | 0.92 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr19_+_36545833 | 0.92 |
ENST00000401500.2
ENST00000270301.7 ENST00000427823.2 |
WDR62
|
WD repeat domain 62 |
| chr1_+_50575292 | 0.92 |
ENST00000371821.1
ENST00000371819.1 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr21_+_39628852 | 0.91 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr18_-_53255766 | 0.89 |
ENST00000566286.1
ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4
|
transcription factor 4 |
| chr1_-_111970353 | 0.87 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr5_-_160973649 | 0.87 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr15_+_79166065 | 0.86 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr1_+_196857144 | 0.84 |
ENST00000367416.2
ENST00000251424.4 ENST00000367418.2 |
CFHR4
|
complement factor H-related 4 |
| chr10_-_52383644 | 0.83 |
ENST00000361781.2
|
SGMS1
|
sphingomyelin synthase 1 |
| chrX_+_49687216 | 0.82 |
ENST00000376088.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr11_+_113779704 | 0.81 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr3_-_52864680 | 0.80 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr19_+_19322758 | 0.78 |
ENST00000252575.6
|
NCAN
|
neurocan |
| chr12_-_52828147 | 0.77 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chrX_-_11284095 | 0.77 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr4_-_110723134 | 0.75 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr1_-_109940550 | 0.75 |
ENST00000256637.6
|
SORT1
|
sortilin 1 |
| chr1_+_100818009 | 0.75 |
ENST00000370125.2
ENST00000361544.6 ENST00000370124.3 |
CDC14A
|
cell division cycle 14A |
| chr17_-_39507064 | 0.74 |
ENST00000007735.3
|
KRT33A
|
keratin 33A |
| chr7_-_50633078 | 0.73 |
ENST00000444124.2
|
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr16_+_81348528 | 0.73 |
ENST00000568107.2
|
GAN
|
gigaxonin |
| chr15_-_58571445 | 0.72 |
ENST00000558231.1
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chrX_+_100878079 | 0.71 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr3_+_132316081 | 0.71 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr8_-_63998590 | 0.71 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr4_+_128702969 | 0.71 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr19_-_6279932 | 0.71 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr8_-_16050288 | 0.70 |
ENST00000350896.3
|
MSR1
|
macrophage scavenger receptor 1 |
| chr12_-_50677255 | 0.67 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr17_-_39538550 | 0.66 |
ENST00000394001.1
|
KRT34
|
keratin 34 |
| chr17_-_56769382 | 0.65 |
ENST00000240361.8
ENST00000349033.5 ENST00000389934.3 |
TEX14
|
testis expressed 14 |
| chr2_+_192109911 | 0.64 |
ENST00000418908.1
ENST00000339514.4 ENST00000392318.3 |
MYO1B
|
myosin IB |
| chr17_+_29664830 | 0.64 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr4_-_170533723 | 0.64 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr10_-_51371321 | 0.63 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr12_+_121163538 | 0.63 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr11_-_10590118 | 0.62 |
ENST00000529598.1
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr19_+_852291 | 0.61 |
ENST00000263621.1
|
ELANE
|
elastase, neutrophil expressed |
| chr2_+_204732487 | 0.61 |
ENST00000302823.3
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr6_-_167571817 | 0.61 |
ENST00000366834.1
|
GPR31
|
G protein-coupled receptor 31 |
| chr5_-_22853429 | 0.59 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr19_-_48547294 | 0.58 |
ENST00000293255.2
|
CABP5
|
calcium binding protein 5 |
| chr15_-_55881135 | 0.58 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chr22_-_38577731 | 0.58 |
ENST00000335539.3
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr22_-_38577782 | 0.58 |
ENST00000430886.1
ENST00000332509.3 ENST00000447598.2 ENST00000435484.1 ENST00000402064.1 ENST00000436218.1 |
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr8_-_16050214 | 0.57 |
ENST00000262101.5
|
MSR1
|
macrophage scavenger receptor 1 |
| chr11_+_60995849 | 0.57 |
ENST00000537932.1
|
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr5_-_138533973 | 0.55 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr1_-_173176452 | 0.54 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_-_151511911 | 0.53 |
ENST00000392801.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr9_-_98269481 | 0.52 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr1_+_171750776 | 0.50 |
ENST00000458517.1
ENST00000362019.3 ENST00000367737.5 ENST00000361735.3 |
METTL13
|
methyltransferase like 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA13
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 33.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 2.8 | 11.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.8 | 21.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.3 | 3.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.2 | 3.5 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.0 | 3.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.9 | 4.5 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.9 | 2.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.8 | 5.3 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.8 | 2.3 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.7 | 3.7 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.7 | 4.0 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 5.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.6 | 1.8 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.6 | 6.5 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.6 | 4.6 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.6 | 3.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.6 | 1.7 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.6 | 2.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.5 | 1.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.5 | 1.5 | GO:1904479 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.5 | 5.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 3.9 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.4 | 4.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 1.6 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.4 | 1.9 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.4 | 1.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.4 | 1.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.4 | 0.7 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.3 | 1.0 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.3 | 1.0 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 3.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 4.4 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 2.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 1.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.3 | 1.2 | GO:0035720 | signal transduction downstream of smoothened(GO:0007227) intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.3 | 1.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 0.9 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.3 | 1.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 1.4 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.3 | 1.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.3 | 5.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 1.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.2 | 1.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 1.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 0.6 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.2 | 2.5 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.2 | 2.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.7 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.2 | 1.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 6.1 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.2 | 1.8 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.2 | 1.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.6 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.1 | 2.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 6.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.1 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 1.8 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 1.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 0.7 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 5.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.4 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 0.6 | GO:1903238 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.1 | 2.8 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.5 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.1 | 0.7 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.5 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 2.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.5 | GO:0097527 | Fas signaling pathway(GO:0036337) necroptotic signaling pathway(GO:0097527) |
| 0.1 | 1.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 2.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.1 | 0.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.1 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 1.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.7 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 1.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 1.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 1.9 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 1.3 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 1.1 | GO:0000165 | MAPK cascade(GO:0000165) |
| 0.0 | 0.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 1.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 2.7 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 4.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 2.2 | GO:0002292 | T cell differentiation involved in immune response(GO:0002292) |
| 0.0 | 0.8 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 1.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 3.0 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 2.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.7 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 1.6 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 1.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 2.2 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.7 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.4 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 21.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.2 | 13.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 11.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.7 | 3.6 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 3.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 11.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 6.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 1.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 1.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.3 | 20.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 5.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.9 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 6.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.0 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 1.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 1.9 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.1 | 2.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 4.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 3.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.8 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 2.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 2.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 4.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 4.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 2.2 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 1.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 5.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.6 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.9 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.9 | 5.6 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 1.9 | 11.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 1.0 | 13.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.9 | 5.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.8 | 4.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 3.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.6 | 1.8 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.6 | 2.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 4.3 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 1.6 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.5 | 3.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 1.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 6.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 1.8 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.4 | 1.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 6.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.1 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.3 | 1.7 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.3 | 1.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.3 | 1.9 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 6.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 1.6 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.3 | 21.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.3 | 3.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 0.8 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 3.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.5 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.2 | 1.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.2 | 0.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.2 | 1.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 2.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 0.7 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 2.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 2.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 2.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.2 | 0.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 4.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 1.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 1.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 1.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.9 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 1.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.8 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 6.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 1.3 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 10.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.7 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 5.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 3.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 3.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 31.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.4 | 23.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 5.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 13.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 6.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 7.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 5.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 3.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 6.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 21.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 11.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 6.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 4.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 17.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 13.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.2 | 6.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 4.6 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 18.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 3.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.9 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 3.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.5 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.9 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |