Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
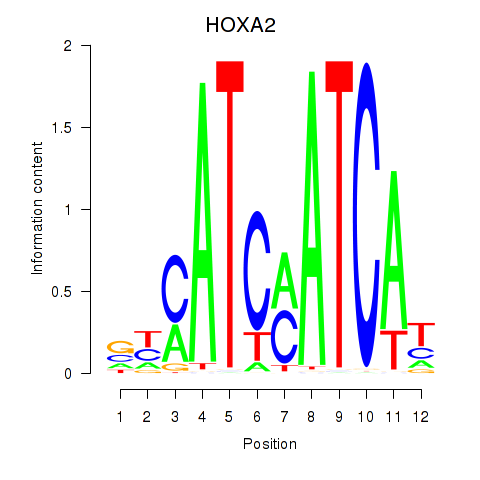
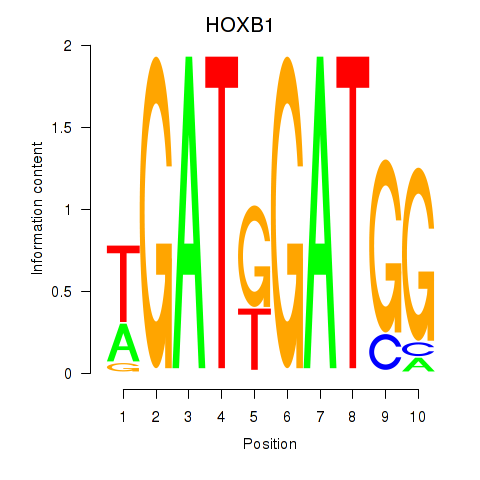
Results for HOXA2_HOXB1
Z-value: 0.25
Transcription factors associated with HOXA2_HOXB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXA2
|
ENSG00000105996.5 | homeobox A2 |
|
HOXB1
|
ENSG00000120094.6 | homeobox B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB1 | hg19_v2_chr17_-_46608272_46608385 | 0.41 | 3.4e-10 | Click! |
| HOXA2 | hg19_v2_chr7_-_27142290_27142430 | 0.26 | 1.1e-04 | Click! |
Activity profile of HOXA2_HOXB1 motif
Sorted Z-values of HOXA2_HOXB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_13835147 | 22.56 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr4_-_90757364 | 17.83 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_111782696 | 16.87 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chrX_-_13835461 | 16.34 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr6_+_151561085 | 15.64 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr14_-_21516590 | 15.43 |
ENST00000555026.1
|
NDRG2
|
NDRG family member 2 |
| chr4_-_90756769 | 14.10 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr6_+_151561506 | 14.10 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_-_111782484 | 14.09 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr7_+_86273218 | 11.07 |
ENST00000361669.2
|
GRM3
|
glutamate receptor, metabotropic 3 |
| chr16_+_15596123 | 7.63 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr12_+_7023491 | 7.27 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr12_+_21679220 | 7.13 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr12_+_101988627 | 7.11 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr3_-_187388173 | 6.79 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr9_-_101471479 | 6.34 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr2_+_12857043 | 6.34 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr16_-_49890016 | 5.89 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr20_+_34742650 | 5.52 |
ENST00000373945.1
ENST00000338074.2 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr19_-_49944806 | 5.37 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr11_-_2162162 | 5.12 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_+_101988774 | 4.96 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr2_+_12857015 | 4.69 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr4_-_102267953 | 4.65 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr18_+_56887381 | 4.62 |
ENST00000256857.2
ENST00000529320.2 ENST00000420468.2 |
GRP
|
gastrin-releasing peptide |
| chr2_-_44588694 | 4.60 |
ENST00000409957.1
|
PREPL
|
prolyl endopeptidase-like |
| chr2_-_44588679 | 4.50 |
ENST00000409411.1
|
PREPL
|
prolyl endopeptidase-like |
| chr12_-_15038779 | 4.22 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr19_-_12997995 | 4.06 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr13_-_110438914 | 4.05 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr6_+_32006042 | 3.77 |
ENST00000418967.2
|
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr5_-_125930929 | 3.75 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chrX_+_82763265 | 3.66 |
ENST00000373200.2
|
POU3F4
|
POU class 3 homeobox 4 |
| chr8_-_143999259 | 3.22 |
ENST00000323110.2
|
CYP11B2
|
cytochrome P450, family 11, subfamily B, polypeptide 2 |
| chr13_+_52586517 | 3.14 |
ENST00000523764.1
ENST00000521508.1 |
ALG11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr6_+_32006159 | 3.00 |
ENST00000478281.1
ENST00000471671.1 ENST00000435122.2 |
CYP21A2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr17_+_37824411 | 2.95 |
ENST00000269582.2
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr12_+_57610562 | 2.94 |
ENST00000349394.5
|
NXPH4
|
neurexophilin 4 |
| chr12_-_49393092 | 2.89 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr8_-_38008783 | 2.86 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr1_-_67390474 | 2.81 |
ENST00000371023.3
ENST00000371022.3 ENST00000371026.3 ENST00000431318.1 |
WDR78
|
WD repeat domain 78 |
| chr12_-_58135903 | 2.76 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr3_+_189507460 | 2.75 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr2_+_68962014 | 2.74 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr1_-_109584716 | 2.64 |
ENST00000531337.1
ENST00000529074.1 ENST00000369965.4 |
WDR47
|
WD repeat domain 47 |
| chrX_+_41583408 | 2.52 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr17_+_37824217 | 2.40 |
ENST00000394246.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr10_+_95848824 | 2.39 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr1_-_109584768 | 2.39 |
ENST00000357672.3
|
WDR47
|
WD repeat domain 47 |
| chrX_-_65253506 | 2.35 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_109584608 | 2.21 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr9_-_73029540 | 2.17 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr7_-_14029515 | 2.17 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr16_-_67427389 | 2.07 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr1_-_223536679 | 2.05 |
ENST00000608996.1
|
SUSD4
|
sushi domain containing 4 |
| chr17_-_46608272 | 2.05 |
ENST00000577092.1
ENST00000239174.6 |
HOXB1
|
homeobox B1 |
| chr20_+_34802295 | 2.02 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chrX_-_47509887 | 1.97 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr9_-_127269661 | 1.93 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr12_+_7282795 | 1.90 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr18_-_53070913 | 1.84 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr10_-_105615164 | 1.78 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr17_+_21729593 | 1.76 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr17_-_37353950 | 1.65 |
ENST00000394310.3
ENST00000394303.3 ENST00000344140.5 |
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr2_-_200323414 | 1.62 |
ENST00000443023.1
|
SATB2
|
SATB homeobox 2 |
| chr11_+_57365150 | 1.59 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_+_89264625 | 1.54 |
ENST00000371996.4
ENST00000371994.4 |
MINPP1
|
multiple inositol-polyphosphate phosphatase 1 |
| chr7_+_79765071 | 1.53 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr6_-_112575838 | 1.50 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr17_+_58755184 | 1.49 |
ENST00000589222.1
ENST00000407086.3 ENST00000390652.5 |
BCAS3
|
breast carcinoma amplified sequence 3 |
| chr8_-_143961236 | 1.48 |
ENST00000377675.3
ENST00000517471.1 ENST00000292427.4 |
CYP11B1
|
cytochrome P450, family 11, subfamily B, polypeptide 1 |
| chr8_+_23386305 | 1.48 |
ENST00000519973.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr6_-_112575687 | 1.42 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr17_+_48609903 | 1.35 |
ENST00000268933.3
|
EPN3
|
epsin 3 |
| chr10_-_79397316 | 1.33 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr7_-_14029283 | 1.32 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr2_-_154335300 | 1.32 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr10_-_98945677 | 1.31 |
ENST00000266058.4
ENST00000371041.3 |
SLIT1
|
slit homolog 1 (Drosophila) |
| chr10_-_79397202 | 1.28 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_-_100995540 | 1.26 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chrX_+_95939711 | 1.17 |
ENST00000373049.4
ENST00000324765.8 |
DIAPH2
|
diaphanous-related formin 2 |
| chr19_+_52074502 | 1.14 |
ENST00000545217.1
ENST00000262259.2 ENST00000596504.1 |
ZNF175
|
zinc finger protein 175 |
| chr8_+_143530791 | 1.02 |
ENST00000517894.1
|
BAI1
|
brain-specific angiogenesis inhibitor 1 |
| chr12_+_57482877 | 1.00 |
ENST00000342556.6
ENST00000357680.4 |
NAB2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr6_-_112575912 | 0.99 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr11_+_68228186 | 0.97 |
ENST00000393799.2
ENST00000393800.2 ENST00000528635.1 ENST00000533127.1 ENST00000529907.1 ENST00000529344.1 ENST00000534534.1 ENST00000524845.1 ENST00000265637.4 ENST00000524904.1 ENST00000393801.3 ENST00000265636.5 ENST00000529710.1 |
PPP6R3
|
protein phosphatase 6, regulatory subunit 3 |
| chr7_-_27170352 | 0.96 |
ENST00000428284.2
ENST00000360046.5 |
HOXA4
|
homeobox A4 |
| chr10_+_18689637 | 0.85 |
ENST00000377315.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr6_+_29426230 | 0.84 |
ENST00000442615.1
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr9_-_26947453 | 0.81 |
ENST00000397292.3
|
PLAA
|
phospholipase A2-activating protein |
| chr17_+_37824700 | 0.81 |
ENST00000581428.1
|
PNMT
|
phenylethanolamine N-methyltransferase |
| chr17_+_39975544 | 0.81 |
ENST00000544340.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr20_+_52105495 | 0.79 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr19_+_47421933 | 0.76 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr2_+_135011731 | 0.75 |
ENST00000281923.2
|
MGAT5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr17_-_42277203 | 0.68 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr3_+_148545586 | 0.68 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr10_-_100995603 | 0.68 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr12_+_21168630 | 0.62 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr15_-_74501310 | 0.60 |
ENST00000423167.2
ENST00000432245.2 |
STRA6
|
stimulated by retinoic acid 6 |
| chr3_+_189507523 | 0.58 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr19_-_41356347 | 0.58 |
ENST00000301141.5
|
CYP2A6
|
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr7_-_27135591 | 0.57 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr1_-_43833628 | 0.56 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr15_-_74501360 | 0.55 |
ENST00000323940.5
|
STRA6
|
stimulated by retinoic acid 6 |
| chrX_+_99899180 | 0.33 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr17_+_39975455 | 0.33 |
ENST00000455106.1
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr6_-_112575758 | 0.31 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr7_+_20370746 | 0.27 |
ENST00000222573.4
|
ITGB8
|
integrin, beta 8 |
| chr19_+_35849723 | 0.26 |
ENST00000594310.1
|
FFAR3
|
free fatty acid receptor 3 |
| chr2_-_200322723 | 0.26 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr1_-_92351769 | 0.23 |
ENST00000212355.4
|
TGFBR3
|
transforming growth factor, beta receptor III |
| chr10_+_102891048 | 0.22 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr4_-_123542224 | 0.19 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr9_+_140135665 | 0.19 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr8_+_23386557 | 0.12 |
ENST00000523930.1
|
SLC25A37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr4_-_5021164 | 0.10 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.4 |
CYTL1
|
cytokine-like 1 |
| chr1_+_178062855 | 0.08 |
ENST00000448150.3
|
RASAL2
|
RAS protein activator like 2 |
| chr14_+_22985251 | 0.03 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA2_HOXB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 70.8 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.2 | 11.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 2.0 | 29.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 1.8 | 5.4 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 1.7 | 31.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.7 | 6.8 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 1.5 | 4.6 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 1.5 | 6.2 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 1.5 | 7.6 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.2 | 4.6 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 1.0 | 15.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.0 | 2.9 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.9 | 12.1 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 3.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.7 | 11.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.7 | 11.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) mineralocorticoid metabolic process(GO:0008212) |
| 0.7 | 2.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.7 | 4.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.6 | 5.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.5 | 3.7 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.5 | 1.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.5 | 1.9 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.5 | 2.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 1.8 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.4 | 1.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 | 0.6 | GO:0021834 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.3 | 2.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.2 | 1.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.2 | 9.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 1.9 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 2.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.2 | 2.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.2 | 1.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 3.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 | 4.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 3.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 2.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.2 | 6.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 1.5 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 5.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 5.4 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.0 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 | 2.3 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 | 0.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.8 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.6 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.1 | 0.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 2.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 4.1 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.0 | 2.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.8 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 1.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 2.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.9 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.2 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 1.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.5 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.1 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 2.8 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.9 | 31.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.2 | 7.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.0 | 31.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 5.4 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.5 | 4.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.5 | 12.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.4 | 2.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 2.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 2.8 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 2.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 11.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 4.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 42.7 | GO:0045121 | membrane raft(GO:0045121) |
| 0.1 | 29.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 15.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.3 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 4.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 8.8 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 3.3 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 5.9 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 25.5 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 1.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 6.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 31.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 2.2 | 11.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 2.1 | 6.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.1 | 6.2 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 1.9 | 29.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 1.6 | 4.7 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 1.3 | 5.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.2 | 7.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 1.0 | 3.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.0 | 31.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.9 | 12.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 11.0 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.5 | 1.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.5 | 1.9 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.5 | 4.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 9.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.4 | 1.5 | GO:0052827 | inositol pentakisphosphate phosphatase activity(GO:0052827) |
| 0.3 | 2.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 3.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 7.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 3.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 9.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 3.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 2.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.1 | 4.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 2.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 6.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 5.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 1.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 9.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 2.9 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 31.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 4.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 2.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 10.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.6 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 12.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 5.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 3.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 3.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 17.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 11.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 31.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 6.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 4.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 4.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 5.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 5.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 12.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 7.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 2.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 5.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 4.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.0 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 6.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 3.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |