Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
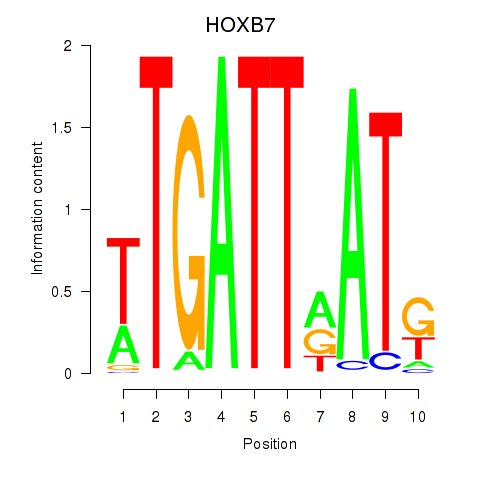
Results for HOXB7
Z-value: 0.58
Transcription factors associated with HOXB7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB7
|
ENSG00000260027.3 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB7 | hg19_v2_chr17_-_46688334_46688385 | -0.10 | 1.4e-01 | Click! |
Activity profile of HOXB7 motif
Sorted Z-values of HOXB7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_20646824 | 13.87 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr10_-_21786179 | 13.48 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr3_-_58613323 | 12.46 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr12_-_91573132 | 12.45 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr18_+_29171689 | 12.00 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr12_-_91546926 | 10.39 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_-_42992856 | 10.15 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr9_-_93405352 | 10.14 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chr2_+_113033164 | 9.89 |
ENST00000409871.1
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr10_+_13142225 | 9.54 |
ENST00000378747.3
|
OPTN
|
optineurin |
| chr3_-_187388173 | 9.07 |
ENST00000287641.3
|
SST
|
somatostatin |
| chr14_+_101299520 | 8.82 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr11_+_57365150 | 8.37 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_+_13142075 | 8.30 |
ENST00000378757.2
ENST00000430081.1 ENST00000378752.3 ENST00000378748.3 |
OPTN
|
optineurin |
| chr12_-_91573249 | 7.98 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr12_+_21679220 | 7.62 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chrX_-_92928557 | 7.44 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr16_+_15596123 | 7.14 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr10_-_15902449 | 6.91 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr19_+_52693259 | 6.89 |
ENST00000322088.6
ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A
|
protein phosphatase 2, regulatory subunit A, alpha |
| chr12_+_20968608 | 6.20 |
ENST00000381541.3
ENST00000540229.1 ENST00000553473.1 ENST00000554957.1 |
LST3
SLCO1B3
SLCO1B7
|
Putative solute carrier organic anion transporter family member 1B7; Uncharacterized protein solute carrier organic anion transporter family, member 1B3 solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr12_+_113229452 | 6.04 |
ENST00000389385.4
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr2_+_234580525 | 5.99 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr8_+_39770803 | 5.96 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr11_-_2162162 | 5.90 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_+_113229737 | 5.85 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr14_+_74417192 | 5.85 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr12_+_113229543 | 5.85 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_-_91573316 | 5.84 |
ENST00000393155.1
|
DCN
|
decorin |
| chr4_-_48014931 | 5.84 |
ENST00000420489.2
ENST00000504722.1 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr9_-_101471479 | 5.70 |
ENST00000259455.2
|
GABBR2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr4_+_113739244 | 5.66 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr12_-_50290839 | 5.56 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr2_+_234580499 | 5.54 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_-_241520525 | 5.30 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr3_+_158787041 | 5.28 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr3_+_189507460 | 5.24 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr12_+_101988627 | 5.20 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr2_-_27886676 | 5.18 |
ENST00000337768.5
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like |
| chr3_+_148545586 | 5.09 |
ENST00000282957.4
ENST00000468341.1 |
CPB1
|
carboxypeptidase B1 (tissue) |
| chr11_-_123525289 | 5.09 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr7_+_77325738 | 5.08 |
ENST00000334955.8
|
RSBN1L
|
round spermatid basic protein 1-like |
| chr3_+_35722487 | 4.96 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_+_72926145 | 4.94 |
ENST00000425662.2
ENST00000453976.2 |
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr13_-_110438914 | 4.90 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr2_-_56150910 | 4.81 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr1_-_67142710 | 4.78 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr14_+_24583836 | 4.73 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr11_+_71927807 | 4.67 |
ENST00000298223.6
ENST00000454954.2 ENST00000541003.1 ENST00000539412.1 ENST00000536778.1 ENST00000535625.1 ENST00000321324.7 |
FOLR2
|
folate receptor 2 (fetal) |
| chr16_-_20339123 | 4.63 |
ENST00000381360.5
|
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr1_+_44584522 | 4.32 |
ENST00000372299.3
|
KLF17
|
Kruppel-like factor 17 |
| chr16_-_21289627 | 4.26 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr14_-_24584138 | 4.23 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr3_+_148415571 | 4.21 |
ENST00000497524.1
ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr15_-_65426174 | 4.19 |
ENST00000204549.4
|
PDCD7
|
programmed cell death 7 |
| chr4_-_100212132 | 4.14 |
ENST00000209668.2
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr14_-_31926701 | 4.11 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr2_+_176972000 | 4.03 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr12_+_101988774 | 4.02 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr9_-_15472730 | 4.00 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr12_+_56401268 | 3.97 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr1_+_196621002 | 3.97 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr6_-_90025011 | 3.96 |
ENST00000402938.3
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr3_+_148508845 | 3.93 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr1_+_19923454 | 3.92 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr16_-_66584059 | 3.90 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr9_-_73029540 | 3.88 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr2_-_228582709 | 3.75 |
ENST00000541617.1
ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr1_+_101702417 | 3.74 |
ENST00000305352.6
|
S1PR1
|
sphingosine-1-phosphate receptor 1 |
| chr21_+_44313375 | 3.74 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr1_+_168148273 | 3.72 |
ENST00000367830.3
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr10_+_13141585 | 3.72 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr5_+_140227357 | 3.72 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr4_+_69962185 | 3.70 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr12_+_52056548 | 3.70 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr22_-_22337146 | 3.69 |
ENST00000398793.2
|
TOP3B
|
topoisomerase (DNA) III beta |
| chr8_+_42128812 | 3.67 |
ENST00000520810.1
ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr1_-_168513229 | 3.67 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2 |
| chr6_-_90024967 | 3.66 |
ENST00000602399.1
|
GABRR2
|
gamma-aminobutyric acid (GABA) A receptor, rho 2 |
| chr19_-_33360647 | 3.63 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr2_+_234600253 | 3.62 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr5_+_140227048 | 3.56 |
ENST00000532602.1
|
PCDHA9
|
protocadherin alpha 9 |
| chr9_-_95166841 | 3.54 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr16_-_20338748 | 3.53 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr8_+_104892639 | 3.52 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr14_+_88851874 | 3.50 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr17_-_26694979 | 3.50 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr4_+_69962212 | 3.48 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr5_+_125695805 | 3.47 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_104893863 | 3.35 |
ENST00000260315.3
ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5
|
caspase 5, apoptosis-related cysteine peptidase |
| chr3_+_159557637 | 3.30 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr6_+_45390222 | 3.30 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr17_-_26695013 | 3.30 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr12_+_81471816 | 3.29 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chr2_+_171036635 | 3.27 |
ENST00000484338.2
ENST00000334231.6 |
MYO3B
|
myosin IIIB |
| chr16_-_66583701 | 3.24 |
ENST00000527800.1
ENST00000525974.1 ENST00000563369.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr4_+_69681710 | 3.21 |
ENST00000265403.7
ENST00000458688.2 |
UGT2B10
|
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr2_+_234637754 | 3.17 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr4_-_141348789 | 3.16 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr3_-_46608010 | 3.15 |
ENST00000395905.3
|
LRRC2
|
leucine rich repeat containing 2 |
| chr12_+_21168630 | 3.14 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr6_+_28317685 | 3.12 |
ENST00000252211.2
ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr9_+_104161123 | 3.12 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chr17_-_64225508 | 3.11 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr18_-_51750948 | 3.09 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr6_-_22297730 | 3.07 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr13_-_33760216 | 3.04 |
ENST00000255486.4
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr3_-_108836977 | 3.00 |
ENST00000232603.5
|
MORC1
|
MORC family CW-type zinc finger 1 |
| chr22_-_22337154 | 2.98 |
ENST00000413067.2
ENST00000437929.1 ENST00000456075.1 ENST00000434517.1 ENST00000424393.1 ENST00000449704.1 ENST00000437103.1 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr16_-_18462221 | 2.98 |
ENST00000528301.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chrX_+_57618269 | 2.97 |
ENST00000374888.1
|
ZXDB
|
zinc finger, X-linked, duplicated B |
| chr10_+_118305435 | 2.95 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr17_+_21030260 | 2.95 |
ENST00000579303.1
|
DHRS7B
|
dehydrogenase/reductase (SDR family) member 7B |
| chr1_+_119957554 | 2.91 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr8_-_18744528 | 2.90 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr19_+_48774586 | 2.88 |
ENST00000594024.1
ENST00000595408.1 ENST00000315849.1 |
ZNF114
|
zinc finger protein 114 |
| chr8_-_110986918 | 2.88 |
ENST00000297404.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr2_+_166095898 | 2.85 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr18_+_22040593 | 2.85 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr1_+_104198377 | 2.84 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr10_-_62332357 | 2.82 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr11_+_6947647 | 2.81 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr19_+_15852203 | 2.80 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chr11_+_89764274 | 2.77 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr4_+_70146217 | 2.74 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr8_+_12803176 | 2.69 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr7_+_119913688 | 2.69 |
ENST00000331113.4
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr15_+_58724184 | 2.68 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr1_-_104238912 | 2.66 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr9_-_127269661 | 2.65 |
ENST00000373588.4
|
NR5A1
|
nuclear receptor subfamily 5, group A, member 1 |
| chr9_-_32526299 | 2.65 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr4_-_70626430 | 2.64 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_44281043 | 2.63 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr19_-_51920952 | 2.63 |
ENST00000356298.5
ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10
|
sialic acid binding Ig-like lectin 10 |
| chr11_-_89541743 | 2.60 |
ENST00000329758.1
|
TRIM49
|
tripartite motif containing 49 |
| chr5_+_54455946 | 2.56 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr10_+_95517660 | 2.55 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr2_+_103035102 | 2.54 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr10_+_13141441 | 2.52 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr4_-_70080449 | 2.52 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr9_-_95166884 | 2.52 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr5_+_140772381 | 2.51 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr22_-_22337204 | 2.50 |
ENST00000430142.1
ENST00000357179.5 |
TOP3B
|
topoisomerase (DNA) III beta |
| chr1_+_117963209 | 2.48 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr11_+_96123158 | 2.43 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr18_+_22040620 | 2.43 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr17_+_41363854 | 2.42 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr4_+_71600144 | 2.41 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr14_-_94789663 | 2.41 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr12_+_59989791 | 2.40 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr18_+_61554932 | 2.39 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr7_-_120498357 | 2.38 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr11_-_55703876 | 2.36 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor, family 5, subfamily I, member 1 |
| chr4_+_88720698 | 2.36 |
ENST00000226284.5
|
IBSP
|
integrin-binding sialoprotein |
| chr3_-_156272924 | 2.34 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr1_-_53608289 | 2.32 |
ENST00000371491.4
|
SLC1A7
|
solute carrier family 1 (glutamate transporter), member 7 |
| chr3_-_169587621 | 2.30 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr6_+_27925019 | 2.30 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr19_+_50016610 | 2.29 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr4_-_69536346 | 2.28 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr8_-_91095099 | 2.28 |
ENST00000265431.3
|
CALB1
|
calbindin 1, 28kDa |
| chr17_-_4938712 | 2.26 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr11_+_28129795 | 2.23 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr2_+_234590556 | 2.21 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr5_+_125758813 | 2.20 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr9_-_27529726 | 2.19 |
ENST00000262244.5
|
MOB3B
|
MOB kinase activator 3B |
| chr3_+_189349162 | 2.18 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr7_+_2636522 | 2.17 |
ENST00000423196.1
|
IQCE
|
IQ motif containing E |
| chr1_-_104239076 | 2.14 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr3_+_181429704 | 2.13 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr12_+_54402790 | 2.12 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr2_+_234621551 | 2.11 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_-_70780770 | 2.11 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr16_-_88729473 | 2.09 |
ENST00000301012.3
ENST00000569177.1 |
MVD
|
mevalonate (diphospho) decarboxylase |
| chr14_+_22337014 | 2.08 |
ENST00000390436.2
|
TRAV13-1
|
T cell receptor alpha variable 13-1 |
| chr5_-_75919253 | 2.07 |
ENST00000296641.4
|
F2RL2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr11_-_63376013 | 2.06 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr1_+_32379174 | 2.05 |
ENST00000391369.1
|
AL136115.1
|
HCG2032337; PRO1848; Uncharacterized protein |
| chr5_+_161112563 | 2.03 |
ENST00000274545.5
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chr12_-_71031185 | 2.02 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr18_+_580367 | 2.02 |
ENST00000327228.3
|
CETN1
|
centrin, EF-hand protein, 1 |
| chr11_+_113779704 | 2.02 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr5_-_125930929 | 2.01 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr5_-_58295712 | 1.99 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr4_+_100737954 | 1.98 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr5_-_132073210 | 1.97 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr19_+_19030478 | 1.96 |
ENST00000247003.4
|
DDX49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr5_+_125758865 | 1.94 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_+_55744166 | 1.92 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr10_-_98273668 | 1.90 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr6_+_29429217 | 1.90 |
ENST00000396792.2
|
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr12_-_71031220 | 1.87 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr9_+_131266963 | 1.87 |
ENST00000309971.4
ENST00000372770.4 |
GLE1
|
GLE1 RNA export mediator |
| chr2_+_157330081 | 1.84 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chrX_+_120181457 | 1.81 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr6_+_25754927 | 1.78 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr13_-_113242439 | 1.78 |
ENST00000375669.3
ENST00000261965.3 |
TUBGCP3
|
tubulin, gamma complex associated protein 3 |
| chr16_-_20364122 | 1.77 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr5_+_161112754 | 1.77 |
ENST00000523217.1
|
GABRA6
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| chrX_+_30248553 | 1.76 |
ENST00000361644.2
|
MAGEB3
|
melanoma antigen family B, 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 36.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 3.0 | 24.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 2.9 | 11.5 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 2.5 | 12.4 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 2.2 | 8.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 1.9 | 5.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.8 | 9.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 1.8 | 7.1 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.7 | 6.9 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.6 | 1.6 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 1.5 | 6.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 1.4 | 7.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 1.3 | 16.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.2 | 3.7 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 1.2 | 3.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 1.2 | 3.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.2 | 4.7 | GO:0071231 | thymidylate synthase biosynthetic process(GO:0050757) regulation of thymidylate synthase biosynthetic process(GO:0050758) cellular response to folic acid(GO:0071231) |
| 1.1 | 1.1 | GO:0035711 | T-helper 1 cell activation(GO:0035711) |
| 1.1 | 4.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 1.1 | 10.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 1.1 | 4.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.9 | 3.7 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.9 | 2.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.8 | 4.9 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.7 | 5.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.7 | 2.8 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.7 | 2.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.7 | 3.4 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.7 | 16.9 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.7 | 2.6 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.7 | 2.6 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.7 | 9.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.7 | 5.9 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 8.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.6 | 26.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 | 3.6 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.6 | 3.0 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.6 | 2.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.6 | 3.9 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.6 | 1.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 9.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.5 | 4.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.5 | 1.6 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.5 | 3.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.5 | 1.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.5 | 1.5 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.5 | 3.3 | GO:0070269 | ectopic germ cell programmed cell death(GO:0035234) pyroptosis(GO:0070269) |
| 0.5 | 1.9 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.5 | 1.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.5 | 2.3 | GO:0072025 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) |
| 0.4 | 4.0 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 1.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.4 | 1.3 | GO:0090294 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.4 | 1.3 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.4 | 1.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.4 | 2.5 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.4 | 3.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 17.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.4 | 3.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.4 | 3.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.4 | 5.6 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.4 | 1.8 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.4 | 7.0 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 3.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 2.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.4 | 3.5 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.4 | 3.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.3 | 3.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.3 | 1.7 | GO:1902617 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) response to fluoride(GO:1902617) |
| 0.3 | 3.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 2.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.3 | 1.0 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 5.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.3 | 2.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 2.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) regulation of adipose tissue development(GO:1904177) |
| 0.3 | 2.0 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.3 | 2.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.3 | 3.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 9.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 2.7 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.3 | 1.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.3 | 0.5 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.3 | 1.8 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 1.2 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 1.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.2 | 1.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 4.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 2.9 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 1.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.2 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 10.9 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.2 | 3.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.2 | 3.0 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.2 | 2.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 3.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 3.7 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 8.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.2 | 1.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 5.3 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.2 | 1.7 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 1.3 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 0.5 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.2 | 0.7 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.2 | 0.7 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.2 | 1.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.2 | 0.9 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.2 | 2.7 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 0.5 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.2 | 6.0 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.2 | 6.1 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.2 | 4.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.5 | GO:0035993 | subthalamic nucleus development(GO:0021763) deltoid tuberosity development(GO:0035993) prolactin secreting cell differentiation(GO:0060127) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 1.9 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 1.2 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 5.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 2.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.6 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 2.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 5.3 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.5 | GO:0060708 | histone H3-K27 acetylation(GO:0043974) leukemia inhibitory factor signaling pathway(GO:0048861) spongiotrophoblast differentiation(GO:0060708) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 1.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 2.3 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 3.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 2.1 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.1 | 8.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 3.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 1.3 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 2.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.7 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.8 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 1.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 2.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 0.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 2.4 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 3.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.9 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 1.1 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.1 | 3.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 1.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 1.5 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 3.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 3.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 9.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 2.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 1.6 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 1.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 3.6 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 1.6 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.7 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 1.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.1 | GO:0070167 | regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 1.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.0 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 4.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 4.0 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 2.6 | GO:1990748 | cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 2.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 4.5 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 0.2 | GO:1903799 | negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 | 1.1 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 4.0 | GO:0051384 | response to glucocorticoid(GO:0051384) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.4 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 2.1 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 1.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 3.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 2.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 1.4 | GO:0048469 | cell maturation(GO:0048469) |
| 0.0 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 2.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 2.0 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.4 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 36.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.9 | 5.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 1.5 | 6.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.9 | 3.5 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.9 | 3.5 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.7 | 11.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.7 | 12.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.6 | 3.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.6 | 3.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 3.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) |
| 0.5 | 1.4 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 1.6 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.4 | 5.8 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.4 | 1.8 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.3 | 9.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 11.4 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 0.9 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 0.9 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.3 | 1.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 3.7 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.3 | 4.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.3 | 4.7 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.3 | 22.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 1.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 17.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 13.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 6.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 2.8 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 8.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 1.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 2.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.2 | 1.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 3.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 5.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 1.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 4.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 5.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 15.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 2.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 5.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 2.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 4.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 9.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.6 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 2.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 8.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 8.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 3.6 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 3.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.2 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 4.9 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.9 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 7.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 4.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 5.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 5.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 6.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 28.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0030286 | cytoplasmic dynein complex(GO:0005868) dynein complex(GO:0030286) |
| 0.0 | 2.8 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.4 | 7.1 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 1.9 | 5.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.8 | 3.7 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.6 | 17.7 | GO:0008430 | selenium binding(GO:0008430) |
| 1.5 | 9.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.5 | 6.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.4 | 4.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 1.4 | 10.8 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.2 | 3.7 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 1.2 | 24.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 1.2 | 45.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.2 | 4.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.0 | 3.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.0 | 5.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.0 | 2.9 | GO:0004769 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 1.0 | 4.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 2.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.8 | 3.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.8 | 2.4 | GO:0005549 | odorant binding(GO:0005549) |
| 0.7 | 2.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.7 | 8.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.7 | 9.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.6 | 3.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.6 | 3.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.6 | 5.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 5.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.6 | 2.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.6 | 2.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.6 | 3.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 3.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.5 | 40.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 2.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 2.0 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.5 | 1.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.5 | 1.8 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.5 | 4.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.4 | 5.8 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.4 | 1.3 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.4 | 1.3 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.4 | 1.3 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.4 | 2.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.4 | 2.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.4 | 2.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.4 | 6.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 1.5 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 8.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.4 | 4.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 3.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.3 | 2.7 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 1.3 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.0 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 11.4 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.3 | 3.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.3 | 9.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.3 | 7.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 0.5 | GO:0032452 | demethylase activity(GO:0032451) histone demethylase activity(GO:0032452) |
| 0.2 | 5.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 3.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 0.7 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.2 | 12.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 4.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 4.0 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.2 | 2.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 3.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 1.6 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.2 | 10.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 0.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 4.0 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 3.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 1.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 0.5 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.2 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 3.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 14.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 4.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 2.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.4 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 5.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 9.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 7.0 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.2 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.1 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 2.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 4.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 3.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 2.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.9 | GO:0030884 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.1 | 1.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.9 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 3.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.9 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.1 | 3.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 6.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 4.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 9.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 3.6 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 3.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 3.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.9 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 3.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.2 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 3.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 2.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 1.2 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 43.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 6.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 3.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 18.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 6.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 3.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 4.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 8.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 9.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 5.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 6.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 3.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 4.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 7.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 17.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 12.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 3.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 42.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 1.4 | 36.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.7 | 8.0 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.5 | 7.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 3.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 14.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 3.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 6.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 8.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.4 | 6.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 13.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.3 | 5.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 5.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 13.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.3 | 7.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 7.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 4.9 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 4.9 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.2 | 10.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 2.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 6.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 3.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 7.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 6.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 5.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 11.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.3 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 3.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 20.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.1 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 6.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 8.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 5.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 6.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 3.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 3.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 7.8 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 2.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |