Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
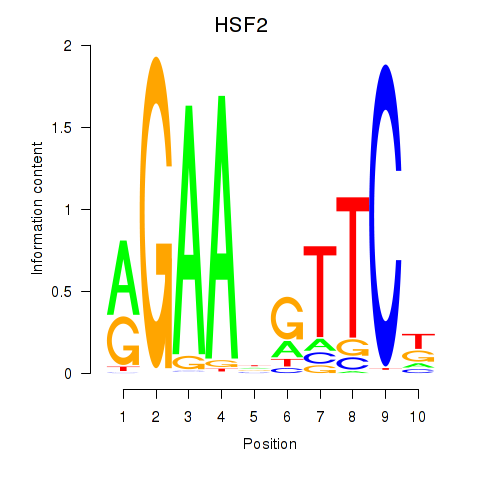
Results for HSF2
Z-value: 0.61
Transcription factors associated with HSF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF2
|
ENSG00000025156.8 | heat shock transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF2 | hg19_v2_chr6_+_122720681_122720713 | -0.15 | 2.5e-02 | Click! |
Activity profile of HSF2 motif
Sorted Z-values of HSF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_75931861 | 28.81 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr12_+_69979210 | 19.59 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr12_+_69979113 | 16.05 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr12_+_69979446 | 15.13 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr18_+_3449695 | 14.54 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_+_33025209 | 13.96 |
ENST00000330899.4
ENST00000544625.1 |
DNAJA1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr2_+_198365122 | 13.54 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr18_-_812231 | 13.19 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr18_+_3449821 | 12.92 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_+_65770227 | 12.20 |
ENST00000527348.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr14_-_102553371 | 12.02 |
ENST00000553585.1
ENST00000216281.8 |
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr12_+_2904102 | 11.93 |
ENST00000001008.4
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr2_+_198365095 | 11.46 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr18_+_3451584 | 11.43 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_+_91926103 | 11.29 |
ENST00000314355.6
|
CKS2
|
CDC28 protein kinase regulatory subunit 2 |
| chr21_-_30445886 | 10.92 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr12_-_53012343 | 10.74 |
ENST00000305748.3
|
KRT73
|
keratin 73 |
| chr2_-_10978103 | 10.58 |
ENST00000404824.2
|
PDIA6
|
protein disulfide isomerase family A, member 6 |
| chr11_+_75273101 | 10.34 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chrX_-_102942961 | 10.08 |
ENST00000434230.1
ENST00000418819.1 ENST00000360458.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr18_+_3451646 | 10.03 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_-_102943022 | 9.71 |
ENST00000433176.2
|
MORF4L2
|
mortality factor 4 like 2 |
| chrX_-_102941596 | 9.59 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr11_-_82997013 | 9.28 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr11_-_82997371 | 9.25 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr11_+_75273246 | 9.20 |
ENST00000526397.1
ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chrX_-_152989531 | 8.80 |
ENST00000458587.2
ENST00000416815.1 |
BCAP31
|
B-cell receptor-associated protein 31 |
| chr11_+_65769550 | 8.77 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chr11_+_65769946 | 8.55 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr7_+_130126165 | 8.39 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr7_-_26240357 | 8.03 |
ENST00000354667.4
ENST00000356674.7 |
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr7_+_26240776 | 7.85 |
ENST00000337620.4
|
CBX3
|
chromobox homolog 3 |
| chr6_+_151646800 | 7.77 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr8_-_141931287 | 7.74 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr11_-_82997420 | 7.70 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr7_+_130126012 | 7.59 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr8_-_81083341 | 7.36 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr15_+_96869165 | 6.87 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr19_-_14628645 | 6.86 |
ENST00000598235.1
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_-_53213780 | 6.83 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr19_-_14629224 | 6.43 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr2_-_188419200 | 6.30 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_-_178128250 | 6.15 |
ENST00000448782.1
ENST00000446151.2 |
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr8_-_81083890 | 5.98 |
ENST00000518937.1
|
TPD52
|
tumor protein D52 |
| chrX_-_153775426 | 5.73 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr11_-_122933043 | 5.68 |
ENST00000534624.1
ENST00000453788.2 ENST00000527387.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr19_+_17416609 | 5.45 |
ENST00000602206.1
|
MRPL34
|
mitochondrial ribosomal protein L34 |
| chr1_-_6453426 | 5.36 |
ENST00000545482.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr4_-_74088800 | 5.17 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr11_-_122932730 | 5.07 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr14_+_89060749 | 5.01 |
ENST00000555900.1
ENST00000406216.3 ENST00000557737.1 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr1_+_206516200 | 5.01 |
ENST00000295713.5
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr3_+_158787041 | 5.00 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr11_-_89956461 | 5.00 |
ENST00000320585.6
|
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr11_-_89956227 | 4.85 |
ENST00000457199.2
ENST00000530765.1 |
CHORDC1
|
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr16_+_765092 | 4.66 |
ENST00000568223.2
|
METRN
|
meteorin, glial cell differentiation regulator |
| chr14_+_89060739 | 4.56 |
ENST00000318308.6
|
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr4_-_22517620 | 4.48 |
ENST00000502482.1
ENST00000334304.5 |
GPR125
|
G protein-coupled receptor 125 |
| chr2_-_219151487 | 4.42 |
ENST00000444881.1
|
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr5_-_146833222 | 4.37 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_123411191 | 4.37 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr13_-_31736027 | 4.35 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr6_+_44215603 | 4.30 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr5_-_146833485 | 4.29 |
ENST00000398514.3
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_+_42651691 | 4.13 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr6_+_149539767 | 3.98 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr14_-_67859422 | 3.79 |
ENST00000556532.1
|
PLEK2
|
pleckstrin 2 |
| chr4_-_159644507 | 3.78 |
ENST00000307720.3
|
PPID
|
peptidylprolyl isomerase D |
| chr2_-_188419078 | 3.62 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_-_6453399 | 3.47 |
ENST00000608083.1
|
ACOT7
|
acyl-CoA thioesterase 7 |
| chr1_+_27248203 | 3.24 |
ENST00000321265.5
|
NUDC
|
nudC nuclear distribution protein |
| chr2_+_11674213 | 3.20 |
ENST00000381486.2
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chrX_+_135230712 | 3.18 |
ENST00000535737.1
|
FHL1
|
four and a half LIM domains 1 |
| chr11_+_86013253 | 3.14 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chr17_+_73663402 | 3.04 |
ENST00000355423.3
|
SAP30BP
|
SAP30 binding protein |
| chrX_+_135279179 | 2.99 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135278908 | 2.92 |
ENST00000539015.1
ENST00000370683.1 |
FHL1
|
four and a half LIM domains 1 |
| chr3_-_185655795 | 2.88 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr12_+_119616447 | 2.58 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr11_+_27062860 | 2.55 |
ENST00000528583.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_-_56494713 | 2.31 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr7_-_150652924 | 2.22 |
ENST00000330883.4
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr2_-_85581701 | 2.19 |
ENST00000295802.4
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr20_+_4666882 | 2.14 |
ENST00000379440.4
ENST00000430350.2 |
PRNP
|
prion protein |
| chr8_-_81083731 | 2.14 |
ENST00000379096.5
|
TPD52
|
tumor protein D52 |
| chr13_+_49684445 | 2.09 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr15_-_43785303 | 2.04 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr6_+_31783291 | 1.81 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr11_+_27062502 | 1.78 |
ENST00000263182.3
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_-_56494882 | 1.73 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr12_-_123011536 | 1.72 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr14_-_25479811 | 1.72 |
ENST00000550887.1
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr6_+_31795506 | 1.64 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr14_-_65409502 | 1.61 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr17_-_56494908 | 1.60 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr18_-_19284724 | 1.53 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr10_+_104535994 | 1.52 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr16_-_30457048 | 1.46 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr2_-_85581623 | 1.42 |
ENST00000449375.1
ENST00000409984.2 ENST00000457495.2 ENST00000263854.6 |
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr13_-_31736478 | 1.34 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr3_-_48632593 | 1.02 |
ENST00000454817.1
ENST00000328333.8 |
COL7A1
|
collagen, type VII, alpha 1 |
| chr6_+_32121789 | 0.99 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr6_+_152126790 | 0.94 |
ENST00000456483.2
|
ESR1
|
estrogen receptor 1 |
| chr4_+_184427235 | 0.89 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr17_-_39661849 | 0.85 |
ENST00000246635.3
ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13
|
keratin 13 |
| chr20_+_57427765 | 0.80 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr6_-_155635583 | 0.75 |
ENST00000367166.4
|
TFB1M
|
transcription factor B1, mitochondrial |
| chr17_-_7297833 | 0.73 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr4_-_151936416 | 0.70 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr14_-_65409438 | 0.65 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr2_-_97405775 | 0.56 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr7_+_120629653 | 0.56 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_+_32121908 | 0.55 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chrX_-_110655391 | 0.42 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr6_-_22297730 | 0.41 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr16_+_71392616 | 0.36 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr2_+_11682790 | 0.23 |
ENST00000389825.3
ENST00000381483.2 |
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr11_-_414948 | 0.19 |
ENST00000530494.1
ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr3_-_129407535 | 0.13 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr20_+_61867235 | 0.13 |
ENST00000342412.6
ENST00000217169.3 |
BIRC7
|
baculoviral IAP repeat containing 7 |
| chr11_-_130298888 | 0.13 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr1_-_216596738 | 0.06 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr5_-_135701164 | 0.05 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr1_-_168698433 | 0.05 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr11_+_66276550 | 0.04 |
ENST00000419755.3
|
CTD-3074O7.11
|
Bardet-Biedl syndrome 1 protein |
| chr1_+_168195229 | 0.01 |
ENST00000271375.4
ENST00000367825.3 |
SFT2D2
|
SFT2 domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.9 | 50.8 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 7.2 | 28.8 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 3.3 | 9.8 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 3.0 | 12.0 | GO:0043335 | protein unfolding(GO:0043335) |
| 2.5 | 29.5 | GO:0015074 | DNA integration(GO:0015074) |
| 2.4 | 19.5 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 2.4 | 19.1 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 2.2 | 8.8 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.2 | 10.8 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 2.1 | 24.7 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.9 | 5.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 1.8 | 8.8 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 1.7 | 6.9 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.6 | 8.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 1.5 | 6.1 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 1.4 | 5.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 1.3 | 5.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.2 | 9.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 1.1 | 4.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.1 | 10.9 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 1.1 | 4.3 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 1.0 | 29.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.9 | 3.8 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.9 | 3.5 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.9 | 5.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.9 | 7.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.7 | 2.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.7 | 2.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.7 | 2.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.7 | 4.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.6 | 9.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.5 | 4.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 | 11.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.5 | 7.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.5 | 5.0 | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) |
| 0.5 | 1.5 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.4 | 4.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 13.2 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.4 | 8.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 11.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.3 | 2.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 1.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.9 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 9.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 10.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.9 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 11.6 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.9 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 11.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 16.0 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 4.0 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 5.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.1 | 35.2 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 2.6 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 4.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.8 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 1.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 4.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 3.2 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 1.0 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 2.1 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.4 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 58.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 2.0 | 14.0 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 1.7 | 5.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 1.5 | 10.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 1.5 | 8.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 1.1 | 4.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.9 | 29.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.6 | 28.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.6 | 7.8 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.6 | 10.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 11.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 2.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 5.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 11.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 11.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.2 | 4.5 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.2 | 8.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 12.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 20.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 5.4 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 31.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 13.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 8.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 3.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 3.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 3.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 4.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 6.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 37.1 | GO:0044429 | mitochondrial part(GO:0044429) |
| 0.0 | 3.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 23.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 9.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 4.2 | GO:0031975 | organelle envelope(GO:0031967) envelope(GO:0031975) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 38.9 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.6 | 28.8 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 4.0 | 11.9 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 2.8 | 30.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 1.9 | 5.7 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 1.9 | 61.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 1.7 | 48.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.6 | 16.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.6 | 11.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 1.1 | 7.8 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.1 | 4.3 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 1.1 | 8.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.9 | 8.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.9 | 8.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.9 | 4.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.9 | 5.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 2.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 10.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.5 | 7.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.5 | 1.5 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.5 | 7.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.4 | 5.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 13.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 2.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.3 | 12.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.3 | 4.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.3 | 1.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.3 | 30.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 3.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 8.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.2 | 13.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 9.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.2 | 6.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 29.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.2 | 3.8 | GO:0080025 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 5.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 4.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 6.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.9 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 7.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 2.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 5.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 3.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 6.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 4.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 28.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.4 | 53.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 12.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.3 | 17.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.3 | 17.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 14.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 14.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 19.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 2.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 4.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 29.5 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 2.0 | 61.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 1.0 | 48.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.7 | 13.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.7 | 4.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.6 | 8.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.6 | 16.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.5 | 41.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.4 | 7.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 20.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 9.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.3 | 5.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 15.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 10.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 8.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 5.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 7.8 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 6.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 8.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 3.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 4.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |