Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for HSFY2
Z-value: 0.71
Transcription factors associated with HSFY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSFY2
|
ENSG00000169953.11 | heat shock transcription factor Y-linked 2 |
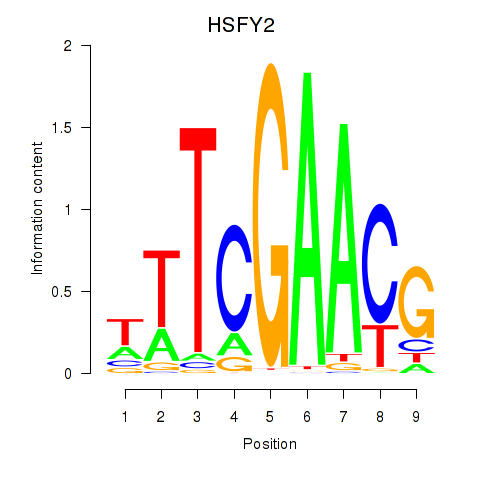
Activity profile of HSFY2 motif
Sorted Z-values of HSFY2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSFY2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.9 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 4.3 | 17.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 4.0 | 31.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 3.8 | 15.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 3.5 | 10.5 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 3.0 | 5.9 | GO:0071029 | nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) |
| 2.9 | 8.6 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 2.8 | 11.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 2.7 | 8.0 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 2.6 | 7.7 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 2.4 | 12.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 2.2 | 13.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 2.1 | 6.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 2.1 | 8.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 2.0 | 5.9 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 1.9 | 5.6 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 1.5 | 5.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.5 | 7.3 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 1.4 | 13.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.4 | 4.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 1.3 | 3.8 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 1.3 | 1.3 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 1.2 | 3.7 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 1.2 | 3.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.1 | 7.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 1.1 | 3.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 1.1 | 14.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 1.1 | 3.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 1.1 | 12.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 1.0 | 4.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.0 | 3.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 1.0 | 3.0 | GO:0035978 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 1.0 | 6.0 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 1.0 | 9.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 1.0 | 5.9 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 1.0 | 9.8 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 1.0 | 7.8 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.9 | 9.9 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.9 | 5.4 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.9 | 3.6 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.9 | 2.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.9 | 3.5 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.8 | 2.5 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.8 | 3.8 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.7 | 3.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.7 | 2.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.7 | 2.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.7 | 3.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.7 | 4.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.7 | 5.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.7 | 2.8 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.7 | 19.9 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.7 | 4.8 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.7 | 6.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.7 | 2.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.7 | 9.8 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 10.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.6 | 9.5 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.6 | 8.0 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.6 | 4.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.6 | 3.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.6 | 4.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.6 | 1.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.6 | 10.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.6 | 18.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.6 | 1.7 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.6 | 12.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.6 | 8.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.6 | 5.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 19.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.5 | 1.5 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.5 | 4.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 8.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.5 | 2.8 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.4 | 3.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.4 | 2.6 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.4 | 1.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.4 | 22.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.4 | 10.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.4 | 3.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 | 7.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 1.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.4 | 1.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.4 | 17.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.4 | 1.6 | GO:0046209 | nitric oxide biosynthetic process(GO:0006809) nitric oxide metabolic process(GO:0046209) |
| 0.4 | 2.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.4 | 18.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.4 | 3.9 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.4 | 2.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 1.5 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.4 | 2.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 1.1 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.3 | 3.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.3 | 11.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 4.5 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.3 | 11.0 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.3 | 1.9 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.3 | 6.7 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.3 | 1.9 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.3 | 11.1 | GO:0045143 | homologous chromosome segregation(GO:0045143) |
| 0.3 | 4.5 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.3 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 0.8 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.3 | 4.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 2.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.3 | 12.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.3 | 2.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 4.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 6.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.2 | 12.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.2 | 1.0 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 7.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.2 | 4.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 4.0 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.2 | 2.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 2.6 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 9.5 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.2 | 0.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.2 | 11.0 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 2.8 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 1.0 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.2 | 3.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 0.8 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.2 | 17.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.2 | 2.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.2 | 6.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.2 | 1.6 | GO:0051964 | netrin-activated signaling pathway(GO:0038007) negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 0.5 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 12.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 1.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.2 | 10.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.2 | 5.1 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 12.4 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) |
| 0.2 | 6.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 2.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 0.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 0.9 | GO:0060501 | negative regulation of keratinocyte differentiation(GO:0045617) positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.2 | 5.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 4.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.1 | 5.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 3.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.8 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.5 | GO:2000232 | regulation of ribosome biogenesis(GO:0090069) regulation of rRNA processing(GO:2000232) |
| 0.1 | 1.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 6.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 8.3 | GO:0090114 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 3.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 2.6 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 1.8 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 2.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 2.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 1.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.9 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.1 | 9.7 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 2.5 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 3.0 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 1.4 | GO:0033561 | regulation of water loss via skin(GO:0033561) establishment of skin barrier(GO:0061436) |
| 0.1 | 3.0 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.1 | 1.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.0 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.7 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 2.0 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 3.8 | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) |
| 0.1 | 1.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.0 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 1.8 | GO:0032986 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 4.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.5 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 5.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.4 | GO:0032259 | methylation(GO:0032259) |
| 0.1 | 3.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 2.8 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 1.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.1 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 1.0 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.3 | GO:0031929 | TOR signaling(GO:0031929) regulation of TOR signaling(GO:0032006) |
| 0.1 | 3.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 4.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 2.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 3.2 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.0 | 3.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 3.2 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 1.9 | GO:0048858 | cell projection morphogenesis(GO:0048858) |
| 0.0 | 0.9 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.4 | GO:0003018 | vascular process in circulatory system(GO:0003018) regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 1.4 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 3.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.8 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.5 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) calcium ion homeostasis(GO:0055074) cellular divalent inorganic cation homeostasis(GO:0072503) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.1 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.7 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.5 | GO:0007369 | gastrulation(GO:0007369) |
| 0.0 | 1.5 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.8 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.6 | GO:0030001 | metal ion transport(GO:0030001) |
| 0.0 | 0.3 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.5 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 1.0 | GO:0034101 | erythrocyte homeostasis(GO:0034101) |
| 0.0 | 1.3 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.5 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 2.6 | 7.8 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 2.3 | 6.9 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 2.3 | 16.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 2.2 | 6.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 2.1 | 14.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 2.1 | 23.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 1.8 | 7.3 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 1.8 | 9.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 1.8 | 10.6 | GO:0001740 | Barr body(GO:0001740) |
| 1.7 | 8.6 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.7 | 13.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 1.6 | 19.7 | GO:0043203 | axon hillock(GO:0043203) |
| 1.6 | 7.8 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 1.5 | 5.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 1.3 | 13.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 1.3 | 6.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.2 | 6.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 1.2 | 5.9 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 1.0 | 3.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 1.0 | 4.9 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.9 | 8.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.9 | 13.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.9 | 4.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.8 | 13.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.8 | 7.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.8 | 9.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.7 | 2.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.7 | 8.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.7 | 12.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.6 | 17.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.6 | 11.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 4.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.5 | 2.2 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.5 | 3.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 12.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.5 | 4.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.5 | 4.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.5 | 9.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 1.5 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.5 | 17.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.4 | 3.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.4 | 2.6 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.4 | 1.3 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.4 | 8.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.4 | 13.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 4.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 8.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 8.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 2.2 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 3.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 2.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.3 | 3.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.3 | 3.0 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 8.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.3 | 19.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 2.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.2 | 2.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 17.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 3.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 4.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 3.5 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 3.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 4.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 7.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 1.0 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.2 | 3.5 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 2.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 9.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 12.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 6.3 | GO:0005657 | replication fork(GO:0005657) |
| 0.2 | 1.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 2.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 1.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 11.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 31.1 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 0.4 | GO:0031090 | organelle membrane(GO:0031090) |
| 0.1 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 7.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.8 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 2.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.2 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.7 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.8 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 61.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 2.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.0 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 6.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 4.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.0 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 24.2 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 4.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 5.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.4 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 20.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 6.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 4.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 19.7 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 3.6 | 10.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 3.5 | 10.5 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 2.8 | 30.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 2.6 | 7.8 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.0 | 6.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 1.9 | 13.5 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 1.9 | 5.6 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 1.8 | 5.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 1.8 | 3.5 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.7 | 9.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 1.6 | 4.7 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 1.4 | 8.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 1.4 | 5.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.1 | 12.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 1.1 | 10.8 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.1 | 4.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.0 | 3.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.9 | 5.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.9 | 41.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.9 | 8.0 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.9 | 8.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.9 | 4.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.9 | 8.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.8 | 3.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.8 | 3.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.7 | 8.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.7 | 2.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.7 | 4.0 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.6 | 2.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.6 | 26.7 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.6 | 8.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.6 | 11.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.5 | 3.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.5 | 2.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 4.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.5 | 3.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.5 | 4.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.5 | 7.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.5 | 7.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 17.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.5 | 4.4 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.5 | 9.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.5 | 4.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.4 | 18.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 11.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 7.8 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.4 | 6.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 4.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 9.0 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.4 | 8.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 7.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.4 | 1.1 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.3 | 4.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 3.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 14.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.3 | 1.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.3 | 9.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 4.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 3.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.3 | 1.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 2.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.7 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 2.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.3 | 8.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.3 | 3.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 3.0 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.2 | 4.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 6.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 9.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 3.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 3.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 7.8 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 4.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 1.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 5.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.8 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 8.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 3.5 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 10.1 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.2 | 2.0 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 5.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 5.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 0.5 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 0.6 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.2 | 0.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 3.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.1 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 2.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 9.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 3.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 3.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 19.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 3.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 24.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.2 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 3.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.7 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 1.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 10.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.7 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 2.4 | GO:0015149 | hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 3.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 7.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 6.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 5.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 10.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 19.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 3.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 1.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) dynein intermediate chain binding(GO:0045505) |
| 0.1 | 2.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 3.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 2.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 4.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 7.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 42.1 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 4.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 7.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.5 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 31.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.5 | 3.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.4 | 22.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 8.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 7.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 18.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 10.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.2 | 2.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 3.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 11.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.2 | 9.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 7.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 6.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 7.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 12.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 7.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 7.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 18.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 1.2 | 13.8 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 1.1 | 18.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.7 | 4.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.7 | 9.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.6 | 9.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 10.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 5.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.5 | 11.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.5 | 29.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.4 | 11.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 11.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 7.8 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 11.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.4 | 17.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.4 | 24.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.4 | 41.7 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.3 | 9.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 7.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 23.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 9.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.3 | 2.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 7.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 9.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.3 | 4.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 8.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 5.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 9.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 3.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 4.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.2 | 4.1 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 4.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 4.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 24.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 10.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 4.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 3.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 7.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 3.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 4.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 6.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 7.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 1.4 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.1 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 7.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 11.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 3.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.6 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 1.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 3.0 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |