Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for IRF6_IRF4_IRF5
Z-value: 0.13
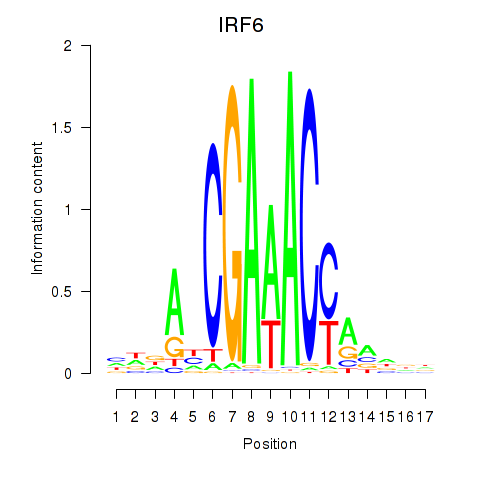
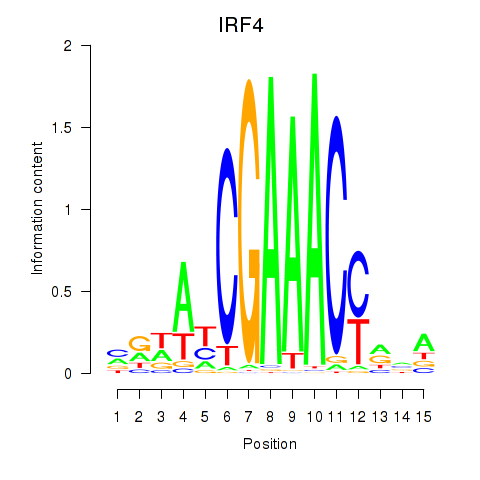
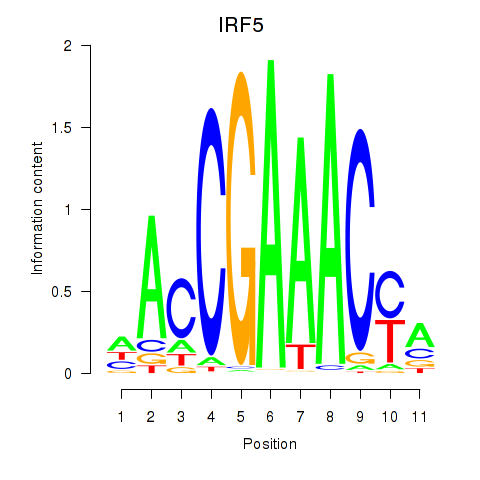
Transcription factors associated with IRF6_IRF4_IRF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRF6
|
ENSG00000117595.6 | interferon regulatory factor 6 |
|
IRF4
|
ENSG00000137265.10 | interferon regulatory factor 4 |
|
IRF5
|
ENSG00000128604.14 | interferon regulatory factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF5 | hg19_v2_chr7_+_128577972_128578047 | 0.18 | 8.9e-03 | Click! |
| IRF6 | hg19_v2_chr1_-_209979375_209979389 | 0.17 | 1.0e-02 | Click! |
| IRF4 | hg19_v2_chr6_+_391739_391759 | 0.12 | 8.7e-02 | Click! |
Activity profile of IRF6_IRF4_IRF5 motif
Sorted Z-values of IRF6_IRF4_IRF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_321050 | 35.16 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_-_615942 | 31.35 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr12_+_113344755 | 30.94 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_146262428 | 30.36 |
ENST00000486631.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_146262488 | 29.85 |
ENST00000487389.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_146262365 | 27.98 |
ENST00000448787.2
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_146262352 | 24.00 |
ENST00000462666.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr3_-_146262637 | 23.14 |
ENST00000472349.1
ENST00000342435.4 |
PLSCR1
|
phospholipid scramblase 1 |
| chr11_-_104817919 | 22.97 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr21_+_42792442 | 22.33 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr3_-_146262293 | 22.26 |
ENST00000448205.1
|
PLSCR1
|
phospholipid scramblase 1 |
| chr1_-_169680745 | 21.93 |
ENST00000236147.4
|
SELL
|
selectin L |
| chr11_-_321340 | 17.93 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr12_+_113344582 | 17.47 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr21_+_42798094 | 17.45 |
ENST00000398598.3
ENST00000455164.2 ENST00000424365.1 |
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr21_+_42733870 | 16.76 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr9_-_32526184 | 15.55 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr17_-_20370847 | 14.22 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr9_-_32526299 | 13.96 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr12_+_113344811 | 13.63 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr11_+_5710919 | 13.18 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr11_-_615570 | 12.23 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr17_+_25958174 | 10.54 |
ENST00000313648.6
ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9
|
lectin, galactoside-binding, soluble, 9 |
| chr7_-_138794081 | 9.23 |
ENST00000464606.1
|
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr22_+_23247030 | 9.13 |
ENST00000390324.2
|
IGLJ3
|
immunoglobulin lambda joining 3 |
| chr7_+_12250833 | 8.94 |
ENST00000396668.3
|
TMEM106B
|
transmembrane protein 106B |
| chr1_-_59043166 | 8.83 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr6_-_82462425 | 8.71 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr17_+_18380051 | 7.84 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr6_+_89791507 | 7.71 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr7_+_12250886 | 7.15 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr17_+_46908350 | 7.08 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chrX_+_49028265 | 6.76 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr3_-_149095652 | 6.55 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr1_+_948803 | 6.20 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr7_+_43798270 | 6.07 |
ENST00000265523.4
ENST00000402924.1 |
BLVRA
|
biliverdin reductase A |
| chr12_+_75874460 | 5.95 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr7_-_138794394 | 5.68 |
ENST00000242351.5
ENST00000471652.1 |
ZC3HAV1
|
zinc finger CCCH-type, antiviral 1 |
| chr2_-_191878874 | 5.66 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr12_+_69004619 | 5.58 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr10_-_91174215 | 5.30 |
ENST00000371837.1
|
LIPA
|
lipase A, lysosomal acid, cholesterol esterase |
| chr12_+_53491220 | 5.25 |
ENST00000548547.1
ENST00000301464.3 |
IGFBP6
|
insulin-like growth factor binding protein 6 |
| chr12_+_113376157 | 5.14 |
ENST00000228928.7
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chrX_+_23801280 | 5.03 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr16_+_28962128 | 5.02 |
ENST00000564978.1
ENST00000320805.4 |
NFATC2IP
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein |
| chr7_-_77045617 | 4.90 |
ENST00000257626.7
|
GSAP
|
gamma-secretase activating protein |
| chr12_+_75874580 | 4.34 |
ENST00000456650.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr13_-_46961580 | 4.33 |
ENST00000378787.3
ENST00000378797.2 ENST00000429979.1 ENST00000378781.3 |
KIAA0226L
|
KIAA0226-like |
| chr12_-_51664058 | 4.27 |
ENST00000605627.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr12_+_75874984 | 4.26 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr7_+_106809406 | 4.06 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr2_-_191878681 | 4.03 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr12_+_62654119 | 4.00 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr14_+_94577074 | 3.87 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr11_+_128563652 | 3.80 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_+_201981527 | 3.47 |
ENST00000441224.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr14_+_22931924 | 3.42 |
ENST00000390477.2
|
TRDC
|
T cell receptor delta constant |
| chr4_-_39529180 | 3.39 |
ENST00000515021.1
ENST00000510490.1 ENST00000316423.6 |
UGDH
|
UDP-glucose 6-dehydrogenase |
| chr2_-_198299726 | 3.38 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chr2_-_89417335 | 3.34 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr1_-_160990886 | 3.13 |
ENST00000537746.1
|
F11R
|
F11 receptor |
| chr19_+_10196781 | 3.13 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr11_-_126138808 | 3.08 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr15_-_85259330 | 3.04 |
ENST00000560266.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr2_-_37384175 | 3.02 |
ENST00000411537.2
ENST00000233057.4 ENST00000395127.2 ENST00000390013.3 |
EIF2AK2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr15_-_85259294 | 2.96 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr11_-_57335280 | 2.96 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr16_-_67970990 | 2.88 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr7_+_94023873 | 2.86 |
ENST00000297268.6
|
COL1A2
|
collagen, type I, alpha 2 |
| chr19_-_17516449 | 2.84 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr2_-_47572105 | 2.80 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr2_+_68961934 | 2.77 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_+_201981119 | 2.75 |
ENST00000395148.2
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr2_+_68961905 | 2.72 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr12_-_51663959 | 2.71 |
ENST00000604188.1
ENST00000398453.3 |
SMAGP
|
small cell adhesion glycoprotein |
| chr17_-_36981556 | 2.66 |
ENST00000536127.1
ENST00000225428.5 |
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr11_-_118134997 | 2.64 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
| chr12_-_51663728 | 2.58 |
ENST00000603864.1
ENST00000605426.1 |
SMAGP
|
small cell adhesion glycoprotein |
| chr1_+_221051699 | 2.57 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr6_+_32605195 | 2.56 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr7_-_148823387 | 2.54 |
ENST00000483014.1
ENST00000378061.2 |
ZNF425
|
zinc finger protein 425 |
| chr9_+_112542591 | 2.44 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr14_+_39736582 | 2.42 |
ENST00000556148.1
ENST00000348007.3 |
CTAGE5
|
CTAGE family, member 5 |
| chr3_-_15374033 | 2.40 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr12_+_121131970 | 2.30 |
ENST00000535656.1
|
MLEC
|
malectin |
| chr19_+_36705504 | 2.27 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr15_-_85259360 | 2.20 |
ENST00000559729.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr9_-_100881466 | 2.13 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr19_-_9546227 | 2.10 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr3_-_15374659 | 2.08 |
ENST00000426925.1
|
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr17_+_6659153 | 2.06 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr4_+_109541772 | 2.05 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr17_-_7164410 | 2.05 |
ENST00000574070.1
|
CLDN7
|
claudin 7 |
| chr18_-_812231 | 1.92 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr17_-_76778339 | 1.91 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr1_-_25558963 | 1.91 |
ENST00000354361.3
|
SYF2
|
SYF2 pre-mRNA-splicing factor |
| chr19_-_9546177 | 1.89 |
ENST00000592292.1
ENST00000588221.1 |
ZNF266
|
zinc finger protein 266 |
| chr2_-_160473114 | 1.88 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr4_+_95373037 | 1.88 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr18_+_3262954 | 1.85 |
ENST00000584539.1
|
MYL12B
|
myosin, light chain 12B, regulatory |
| chr4_-_103748271 | 1.84 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_77134742 | 1.83 |
ENST00000452464.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr16_+_3068393 | 1.82 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr11_+_130029457 | 1.78 |
ENST00000278742.5
|
ST14
|
suppression of tumorigenicity 14 (colon carcinoma) |
| chr13_-_76056250 | 1.78 |
ENST00000377636.3
ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4
|
TBC1 domain family, member 4 |
| chr6_+_64282447 | 1.75 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr18_-_812517 | 1.72 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr16_-_88851618 | 1.71 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr1_-_89357179 | 1.69 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr12_+_70133152 | 1.68 |
ENST00000550536.1
ENST00000362025.5 |
RAB3IP
|
RAB3A interacting protein |
| chr11_+_695614 | 1.67 |
ENST00000608174.1
ENST00000397512.3 |
TMEM80
|
transmembrane protein 80 |
| chr20_-_20033052 | 1.67 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr18_+_42260861 | 1.58 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr7_-_20256965 | 1.55 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr2_+_7017796 | 1.55 |
ENST00000382040.3
|
RSAD2
|
radical S-adenosyl methionine domain containing 2 |
| chr16_+_3062457 | 1.52 |
ENST00000445369.2
|
CLDN9
|
claudin 9 |
| chr12_+_124069070 | 1.47 |
ENST00000262225.3
ENST00000438031.2 |
TMED2
|
transmembrane emp24 domain trafficking protein 2 |
| chr19_+_52264104 | 1.46 |
ENST00000340023.6
|
FPR2
|
formyl peptide receptor 2 |
| chr1_-_54355430 | 1.44 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr19_+_12273866 | 1.41 |
ENST00000425827.1
ENST00000439995.1 ENST00000343979.4 ENST00000398616.2 ENST00000418338.1 |
ZNF136
|
zinc finger protein 136 |
| chr15_+_41136586 | 1.40 |
ENST00000431806.1
|
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr4_-_77135046 | 1.36 |
ENST00000264896.2
|
SCARB2
|
scavenger receptor class B, member 2 |
| chr10_+_91174314 | 1.33 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr1_+_179262905 | 1.31 |
ENST00000539888.1
ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1
|
sterol O-acyltransferase 1 |
| chr2_+_201980827 | 1.29 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr3_-_155572164 | 1.27 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr11_+_809961 | 1.26 |
ENST00000530797.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr3_-_185655795 | 1.25 |
ENST00000342294.4
ENST00000382191.4 ENST00000453386.2 |
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr16_+_30751953 | 1.21 |
ENST00000483578.1
|
RP11-2C24.4
|
RP11-2C24.4 |
| chr2_+_68962014 | 1.20 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr11_+_809647 | 1.20 |
ENST00000321153.4
|
RPLP2
|
ribosomal protein, large, P2 |
| chr6_+_32605134 | 1.18 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr12_+_52643077 | 1.17 |
ENST00000553310.2
ENST00000544024.1 |
KRT86
|
keratin 86 |
| chr16_+_68119324 | 1.15 |
ENST00000349223.5
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr10_+_71561649 | 1.14 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr16_+_12058961 | 1.13 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr17_-_39677971 | 1.11 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr19_+_52264449 | 1.10 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr4_-_89080003 | 1.07 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chrX_-_77041685 | 1.06 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr5_-_61699698 | 1.06 |
ENST00000506390.1
ENST00000199320.4 |
DIMT1
|
DIM1 dimethyladenosine transferase 1 homolog (S. cerevisiae) |
| chr12_-_50616382 | 1.05 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chrX_+_47420516 | 1.03 |
ENST00000377045.4
ENST00000290277.6 ENST00000377039.2 |
ARAF
|
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr12_+_14518598 | 1.03 |
ENST00000261168.4
ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr5_-_95297534 | 1.02 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr5_-_95297678 | 1.02 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr3_-_182880541 | 1.02 |
ENST00000470251.1
ENST00000265598.3 |
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr16_+_68119247 | 1.01 |
ENST00000575270.1
|
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr2_-_55920952 | 1.01 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr1_+_46016703 | 1.01 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr8_-_22926623 | 1.00 |
ENST00000276431.4
|
TNFRSF10B
|
tumor necrosis factor receptor superfamily, member 10b |
| chr19_-_4723761 | 0.99 |
ENST00000597849.1
ENST00000598800.1 ENST00000602161.1 ENST00000597726.1 ENST00000601130.1 ENST00000262960.9 |
DPP9
|
dipeptidyl-peptidase 9 |
| chr16_+_12059050 | 0.95 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr10_+_71562180 | 0.95 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_+_208423840 | 0.94 |
ENST00000539789.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr11_-_66056596 | 0.92 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_+_221054584 | 0.92 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr2_+_85811525 | 0.90 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chr15_-_30113676 | 0.89 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr11_+_32851487 | 0.89 |
ENST00000257836.3
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr2_+_163175394 | 0.88 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr19_-_47616992 | 0.86 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr11_+_126173647 | 0.85 |
ENST00000263579.4
|
DCPS
|
decapping enzyme, scavenger |
| chr16_+_12059091 | 0.83 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr10_-_72648541 | 0.83 |
ENST00000299299.3
|
PCBD1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr10_+_71561704 | 0.81 |
ENST00000520267.1
|
COL13A1
|
collagen, type XIII, alpha 1 |
| chr15_+_90544532 | 0.80 |
ENST00000268154.4
|
ZNF710
|
zinc finger protein 710 |
| chr2_+_99797542 | 0.78 |
ENST00000338148.3
ENST00000512183.2 |
MRPL30
C2orf15
|
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr19_-_50528584 | 0.77 |
ENST00000594092.1
ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3
|
vaccinia related kinase 3 |
| chr2_-_163175133 | 0.76 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr11_-_67141090 | 0.74 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr19_+_10196981 | 0.72 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr10_+_121410882 | 0.71 |
ENST00000369085.3
|
BAG3
|
BCL2-associated athanogene 3 |
| chr19_+_55897297 | 0.70 |
ENST00000431533.2
ENST00000428193.2 ENST00000558815.1 ENST00000560583.1 ENST00000560055.1 ENST00000559463.1 |
RPL28
|
ribosomal protein L28 |
| chr4_-_48782259 | 0.68 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr22_-_31688431 | 0.67 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr6_+_43738444 | 0.66 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr1_+_78769549 | 0.66 |
ENST00000370758.1
|
PTGFR
|
prostaglandin F receptor (FP) |
| chr1_+_236687881 | 0.66 |
ENST00000526634.1
|
LGALS8
|
lectin, galactoside-binding, soluble, 8 |
| chr11_-_66056478 | 0.60 |
ENST00000431556.2
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr3_+_156544057 | 0.58 |
ENST00000498839.1
ENST00000470811.1 ENST00000356539.4 ENST00000483177.1 ENST00000477399.1 ENST00000491763.1 |
LEKR1
|
leucine, glutamate and lysine rich 1 |
| chr1_-_25558984 | 0.56 |
ENST00000236273.4
|
SYF2
|
SYF2 pre-mRNA-splicing factor |
| chr10_+_114710516 | 0.55 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr9_+_112542572 | 0.53 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr2_-_197664366 | 0.51 |
ENST00000409364.3
ENST00000263956.3 |
GTF3C3
|
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr16_+_89642120 | 0.50 |
ENST00000268720.5
ENST00000319518.8 |
CPNE7
|
copine VII |
| chr3_-_46037299 | 0.50 |
ENST00000296137.2
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr2_-_70780572 | 0.45 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chr19_-_52511334 | 0.43 |
ENST00000602063.1
ENST00000597747.1 ENST00000594083.1 ENST00000593650.1 ENST00000599631.1 ENST00000598071.1 ENST00000601178.1 ENST00000376716.5 ENST00000391795.3 |
ZNF615
|
zinc finger protein 615 |
| chr17_+_78965624 | 0.41 |
ENST00000325167.5
|
CHMP6
|
charged multivesicular body protein 6 |
| chr5_+_133707252 | 0.41 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr9_+_97488939 | 0.39 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr3_+_118892362 | 0.36 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr1_+_241695424 | 0.34 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr13_-_113242439 | 0.33 |
ENST00000375669.3
ENST00000261965.3 |
TUBGCP3
|
tubulin, gamma complex associated protein 3 |
| chr6_-_94129244 | 0.33 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr1_+_241695670 | 0.32 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chrX_-_48755030 | 0.32 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr19_+_18942720 | 0.29 |
ENST00000262803.5
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr12_-_56753858 | 0.29 |
ENST00000314128.4
ENST00000557235.1 ENST00000418572.2 |
STAT2
|
signal transducer and activator of transcription 2, 113kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF6_IRF4_IRF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.7 | 157.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 8.7 | 43.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 7.6 | 30.3 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 5.1 | 56.5 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 3.5 | 10.5 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 2.4 | 9.7 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 2.3 | 59.0 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 2.1 | 23.0 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 2.1 | 12.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 1.3 | 7.6 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 1.1 | 8.8 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 1.0 | 71.8 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 1.0 | 3.0 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 1.0 | 4.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 1.0 | 7.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.8 | 3.2 | GO:1905123 | regulation of endosome organization(GO:1904978) regulation of glucosylceramidase activity(GO:1905123) |
| 0.7 | 7.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.6 | 3.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.6 | 1.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 3.4 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.5 | 21.9 | GO:0033198 | response to ATP(GO:0033198) |
| 0.4 | 1.8 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.4 | 1.8 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.4 | 6.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.4 | 1.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.4 | 8.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.4 | 1.5 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.4 | 1.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.3 | 1.7 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.3 | 1.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.3 | 1.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.3 | 1.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.3 | 16.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.3 | 5.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.3 | 2.5 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 3.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 1.3 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 13.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.2 | 0.7 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 0.7 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 4.9 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 2.7 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 3.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 4.6 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.2 | 9.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 0.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 0.3 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 5.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.2 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.9 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.3 | GO:0060948 | cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.1 | 5.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 5.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.1 | 0.7 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 2.1 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.7 | GO:1904977 | lymphatic endothelial cell migration(GO:1904977) |
| 0.1 | 2.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 2.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 3.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.7 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.1 | 2.2 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.1 | 1.9 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.4 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) xenobiotic transport(GO:0042908) |
| 0.1 | 0.8 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 8.7 | GO:0030193 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 0.7 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.5 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.1 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.1 | 1.9 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.8 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.2 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.1 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 4.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.3 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.1 | 1.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 3.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.3 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 2.9 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 4.7 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 3.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 5.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 1.0 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0072178 | nephric duct morphogenesis(GO:0072178) |
| 0.0 | 2.9 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 2.9 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 1.0 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 2.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 9.0 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 2.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 3.7 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.8 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 23.0 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 1.2 | 8.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.8 | 3.0 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.5 | 5.2 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.5 | 68.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.4 | 7.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.4 | 3.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 165.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.4 | 2.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 1.7 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 8.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 1.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 34.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.2 | 3.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 0.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 2.9 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 11.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 6.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 13.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 1.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 3.4 | GO:0005686 | U2 snRNP(GO:0005686) U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 16.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 42.3 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 11.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 2.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.1 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 18.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.3 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 21.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 7.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 1.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 4.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 53.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 3.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 9.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 2.6 | GO:0005856 | cytoskeleton(GO:0005856) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.7 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 14.3 | 157.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 6.0 | 59.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 3.5 | 10.5 | GO:0048030 | disaccharide binding(GO:0048030) |
| 3.1 | 21.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 1.6 | 23.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.5 | 6.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 1.1 | 5.3 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 3.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.9 | 11.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.7 | 4.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.6 | 2.6 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.5 | 11.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.5 | 5.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.5 | 50.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.4 | 1.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 1.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.0 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 30.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.3 | 1.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 3.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 6.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 0.7 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 1.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.8 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.2 | 2.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 6.6 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.2 | 1.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.2 | 54.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 3.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 1.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.9 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 1.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 2.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 2.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 25.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 7.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 5.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 5.0 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.1 | 1.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 2.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 1.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 14.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 14.3 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 3.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 11.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 2.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.7 | 21.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 3.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.3 | 47.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 7.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 22.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 8.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 33.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 1.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 4.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 4.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 4.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 5.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 72.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 2.3 | 189.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.5 | 8.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 22.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.3 | 8.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 2.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 5.6 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 3.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 3.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 21.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 6.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 6.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 13.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.1 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 3.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 3.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 5.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 2.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |