Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for ISL1
Z-value: 0.18
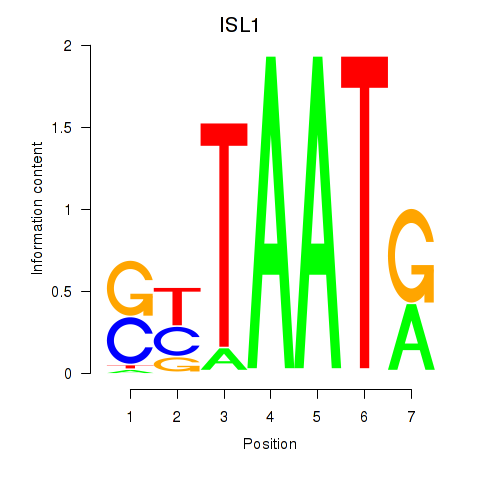
Transcription factors associated with ISL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL1
|
ENSG00000016082.10 | ISL LIM homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL1 | hg19_v2_chr5_+_50678921_50678921 | -0.08 | 2.5e-01 | Click! |
Activity profile of ISL1 motif
Sorted Z-values of ISL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175711133 | 36.26 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr4_-_176923483 | 18.37 |
ENST00000280187.7
ENST00000512509.1 |
GPM6A
|
glycoprotein M6A |
| chr12_-_50290839 | 15.57 |
ENST00000552863.1
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr4_-_186877806 | 13.06 |
ENST00000355634.5
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr14_-_60337684 | 12.33 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr8_+_26371763 | 12.28 |
ENST00000521913.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr1_-_95391315 | 11.81 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr2_-_224467093 | 11.22 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chrX_-_92928557 | 11.14 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr5_+_140772381 | 10.45 |
ENST00000398604.2
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr3_+_158787041 | 10.38 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr4_-_186877502 | 10.26 |
ENST00000431902.1
ENST00000284776.7 ENST00000415274.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr18_-_21977748 | 10.01 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr6_-_116575226 | 9.89 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chrX_+_55478538 | 9.79 |
ENST00000342972.1
|
MAGEH1
|
melanoma antigen family H, 1 |
| chr1_-_156217875 | 9.03 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr3_+_35721106 | 8.82 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_156217822 | 8.22 |
ENST00000368270.1
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr18_-_5544241 | 7.75 |
ENST00000341928.2
ENST00000540638.2 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr8_-_101571964 | 6.93 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr8_+_21911054 | 6.92 |
ENST00000519850.1
ENST00000381470.3 |
DMTN
|
dematin actin binding protein |
| chr9_-_113761720 | 6.39 |
ENST00000541779.1
ENST00000374430.2 |
LPAR1
|
lysophosphatidic acid receptor 1 |
| chrX_+_135252050 | 5.98 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chrX_+_135251783 | 5.89 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chrX_-_102941596 | 5.81 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr9_+_103947311 | 5.76 |
ENST00000395056.2
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr11_-_134281812 | 5.52 |
ENST00000392580.1
ENST00000312527.4 |
B3GAT1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr8_-_101571933 | 5.41 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr4_-_87281196 | 5.39 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_30887948 | 5.01 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr5_+_140248518 | 4.97 |
ENST00000398640.2
|
PCDHA11
|
protocadherin alpha 11 |
| chr4_-_87281224 | 4.96 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_169418195 | 4.95 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr5_-_138210977 | 4.74 |
ENST00000274711.6
ENST00000521094.2 |
LRRTM2
|
leucine rich repeat transmembrane neuronal 2 |
| chr4_+_113970772 | 4.61 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr2_+_166152283 | 4.49 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr2_+_172543919 | 4.47 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chrX_-_23926004 | 4.37 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr3_-_33686743 | 4.24 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_19540266 | 4.24 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_19540086 | 4.21 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr7_+_16793160 | 3.98 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr5_-_142782862 | 3.71 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr18_+_72167096 | 3.67 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr18_-_53177984 | 3.62 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr18_-_52989217 | 3.49 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr7_-_31380502 | 3.43 |
ENST00000297142.3
|
NEUROD6
|
neuronal differentiation 6 |
| chr5_+_137203465 | 3.21 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chrX_+_135251835 | 3.21 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr14_+_20811766 | 3.12 |
ENST00000250416.5
ENST00000527915.1 |
PARP2
|
poly (ADP-ribose) polymerase 2 |
| chr5_+_140593509 | 3.03 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr10_+_123923205 | 3.00 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr10_-_62332357 | 3.00 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr5_-_94417339 | 2.88 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr18_-_52989525 | 2.86 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr5_+_137203557 | 2.69 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr5_+_32788945 | 2.66 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr18_+_46065393 | 2.65 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr7_+_90338712 | 2.48 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr4_+_4861385 | 2.48 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr7_+_129932974 | 2.45 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr3_-_149095652 | 2.43 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr20_+_61448376 | 2.36 |
ENST00000343916.3
|
COL9A3
|
collagen, type IX, alpha 3 |
| chr3_-_192445289 | 2.24 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr11_-_102668879 | 2.21 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr6_+_167704798 | 2.02 |
ENST00000230256.3
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr16_+_2198604 | 2.00 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr15_+_65822756 | 1.99 |
ENST00000562901.1
ENST00000261875.5 ENST00000442729.2 ENST00000565299.1 ENST00000568793.1 |
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr1_-_160254913 | 1.97 |
ENST00000440949.3
ENST00000368072.5 ENST00000608310.1 ENST00000556710.1 |
PEX19
DCAF8
DCAF8
|
peroxisomal biogenesis factor 19 DDB1 and CUL4 associated factor 8 DDB1- and CUL4-associated factor 8 |
| chr5_+_137203541 | 1.93 |
ENST00000421631.2
|
MYOT
|
myotilin |
| chr12_+_14572070 | 1.89 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr19_-_50143452 | 1.88 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr22_+_40390930 | 1.84 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr11_-_18258342 | 1.81 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr6_+_167704838 | 1.77 |
ENST00000366829.2
|
UNC93A
|
unc-93 homolog A (C. elegans) |
| chr10_+_135340859 | 1.76 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr12_-_57037284 | 1.72 |
ENST00000551570.1
|
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr3_+_179322481 | 1.71 |
ENST00000259037.3
|
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_+_82266053 | 1.64 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr15_+_59279851 | 1.59 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr17_-_15501932 | 1.57 |
ENST00000583965.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr10_-_95241951 | 1.54 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr12_-_53012343 | 1.48 |
ENST00000305748.3
|
KRT73
|
keratin 73 |
| chr19_-_17488143 | 1.48 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr11_+_22688150 | 1.46 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr2_+_28618532 | 1.46 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2 |
| chr17_-_64225508 | 1.44 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr6_+_36853607 | 1.40 |
ENST00000480824.2
ENST00000355190.3 ENST00000373685.1 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr10_-_95242044 | 1.40 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr20_+_60174827 | 1.35 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr1_+_100598691 | 1.32 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr6_-_11779014 | 1.31 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr1_+_206972215 | 1.29 |
ENST00000340758.2
|
IL19
|
interleukin 19 |
| chr17_+_66521936 | 1.29 |
ENST00000592800.1
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr3_+_157154578 | 1.27 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr11_-_62783276 | 1.25 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr11_-_62783303 | 1.23 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr1_+_35734562 | 1.19 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr16_+_3493611 | 1.15 |
ENST00000407558.4
ENST00000572169.1 ENST00000572757.1 ENST00000573593.1 ENST00000570372.1 ENST00000424546.2 ENST00000575733.1 ENST00000573201.1 ENST00000574950.1 ENST00000573580.1 ENST00000608722.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chrY_-_6740649 | 1.11 |
ENST00000383036.1
ENST00000383037.4 |
AMELY
|
amelogenin, Y-linked |
| chr2_+_171034646 | 1.09 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr12_-_10251603 | 1.03 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr3_-_119276575 | 1.02 |
ENST00000383669.3
ENST00000383668.3 |
CD80
|
CD80 molecule |
| chr7_-_107880508 | 0.95 |
ENST00000425651.2
|
NRCAM
|
neuronal cell adhesion molecule |
| chr1_-_8000872 | 0.91 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr21_-_47575481 | 0.89 |
ENST00000291670.5
ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD
|
formimidoyltransferase cyclodeaminase |
| chr2_-_214016314 | 0.85 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr6_-_111136513 | 0.85 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr4_+_169418255 | 0.85 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_-_10251576 | 0.83 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr22_-_43355858 | 0.83 |
ENST00000402229.1
ENST00000407585.1 ENST00000453079.1 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr1_-_79472365 | 0.82 |
ENST00000370742.3
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr3_-_138312971 | 0.81 |
ENST00000485115.1
ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70
|
centrosomal protein 70kDa |
| chr16_+_47496023 | 0.79 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr6_+_31515337 | 0.75 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr10_-_17659357 | 0.74 |
ENST00000326961.6
ENST00000361271.3 |
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr9_+_2029019 | 0.73 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_211421262 | 0.72 |
ENST00000233072.5
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr3_+_148508845 | 0.71 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr9_+_27109133 | 0.70 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr12_-_10251539 | 0.69 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr10_-_115613828 | 0.66 |
ENST00000361384.2
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr2_-_217560248 | 0.66 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr17_-_47045949 | 0.61 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr5_+_32710736 | 0.61 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr2_+_234216454 | 0.61 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr8_+_119294456 | 0.60 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr21_-_34185989 | 0.56 |
ENST00000487113.1
ENST00000382373.4 |
C21orf62
|
chromosome 21 open reading frame 62 |
| chr10_-_116444371 | 0.55 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr12_-_121454148 | 0.50 |
ENST00000535367.1
ENST00000538296.1 ENST00000445832.3 ENST00000536407.2 ENST00000366211.2 ENST00000539736.1 ENST00000288757.3 ENST00000537817.1 |
C12orf43
|
chromosome 12 open reading frame 43 |
| chr5_+_34757309 | 0.49 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr8_+_70404996 | 0.47 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr21_-_34186006 | 0.45 |
ENST00000490358.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr18_+_55018044 | 0.45 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr1_+_79086088 | 0.44 |
ENST00000370751.5
ENST00000342282.3 |
IFI44L
|
interferon-induced protein 44-like |
| chr14_-_92198403 | 0.43 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr21_-_42219065 | 0.42 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chrX_+_65382433 | 0.38 |
ENST00000374727.3
|
HEPH
|
hephaestin |
| chr7_-_44580861 | 0.36 |
ENST00000546276.1
ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1
|
NPC1-like 1 |
| chr1_+_221054584 | 0.36 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr18_-_13915530 | 0.36 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr11_-_66496430 | 0.35 |
ENST00000533211.1
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2 |
| chr1_+_197170592 | 0.30 |
ENST00000535699.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr9_+_131084815 | 0.28 |
ENST00000300452.3
ENST00000609948.1 |
COQ4
|
coenzyme Q4 |
| chr2_-_55920952 | 0.27 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chrX_-_100129128 | 0.27 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr16_-_23464459 | 0.26 |
ENST00000307149.5
|
COG7
|
component of oligomeric golgi complex 7 |
| chr17_-_2996290 | 0.26 |
ENST00000331459.1
|
OR1D2
|
olfactory receptor, family 1, subfamily D, member 2 |
| chr12_-_102591604 | 0.26 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr1_-_93257951 | 0.25 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr12_-_28124903 | 0.23 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr8_-_25281747 | 0.23 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr1_+_197382957 | 0.21 |
ENST00000367397.1
|
CRB1
|
crumbs homolog 1 (Drosophila) |
| chr3_+_38307293 | 0.21 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr22_-_31885727 | 0.18 |
ENST00000330125.5
ENST00000344710.5 ENST00000397518.1 |
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr11_+_30252549 | 0.16 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr18_+_46065483 | 0.14 |
ENST00000382998.4
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr11_+_60995849 | 0.14 |
ENST00000537932.1
|
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr3_+_119501557 | 0.13 |
ENST00000337940.4
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr6_+_168418553 | 0.12 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
| chr13_-_108870623 | 0.12 |
ENST00000405925.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr12_-_52761262 | 0.11 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr3_-_167191814 | 0.09 |
ENST00000466903.1
ENST00000264677.4 |
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chrX_-_30327495 | 0.06 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr22_-_32651326 | 0.06 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr2_+_170366203 | 0.03 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr8_-_59412717 | 0.03 |
ENST00000301645.3
|
CYP7A1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chrX_-_100129320 | 0.02 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 1.6 | 6.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.5 | 4.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.3 | 10.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.2 | 6.9 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.1 | 7.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.1 | 36.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 1.0 | 3.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 1.0 | 15.6 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.8 | 2.5 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.7 | 3.0 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.7 | 11.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 23.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.5 | 4.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.5 | 11.8 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.5 | 3.7 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.5 | 18.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 4.5 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 4.7 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.4 | 1.8 | GO:0010193 | response to ozone(GO:0010193) |
| 0.3 | 4.5 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.3 | 0.7 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.3 | 0.9 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 6.1 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 2.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.3 | 2.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.3 | 1.3 | GO:0044793 | negative regulation by host of viral process(GO:0044793) negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 1.4 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.2 | 15.1 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.2 | 0.9 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 | 10.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 1.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.2 | 5.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 2.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 1.4 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.2 | 3.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 5.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.2 | 21.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 5.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 0.9 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 1.0 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.7 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 14.1 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.1 | 0.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 4.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 18.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.6 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 2.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.3 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 1.3 | GO:2000480 | negative regulation of activated T cell proliferation(GO:0046007) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.3 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 11.9 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.1 | 1.5 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.3 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 2.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 2.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 0.4 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 15.0 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.7 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 8.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 2.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 1.8 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 4.0 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 2.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 3.0 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.2 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.8 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 2.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.4 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 3.3 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.5 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.1 | GO:0046606 | negative regulation of centrosome cycle(GO:0046606) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.8 | 2.5 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.7 | 12.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.6 | 4.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.6 | 11.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.5 | 9.9 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.5 | 2.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.4 | 4.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.7 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 8.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 38.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 5.8 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 4.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 6.4 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 1.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 11.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 18.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 2.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 8.8 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.1 | 5.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.0 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 6.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 5.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 8.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 11.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 9.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 9.7 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 2.8 | 8.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 1.8 | 5.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.7 | 10.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.2 | 3.7 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 1.0 | 3.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.9 | 23.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.8 | 36.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.8 | 10.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 3.7 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.7 | 6.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.5 | 1.6 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.5 | 11.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 4.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.3 | 14.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 13.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 1.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 10.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.3 | 13.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 2.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 4.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.7 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 4.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 4.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 17.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.1 | 18.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 17.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.8 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 4.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 2.5 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 1.3 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 3.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.9 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 1.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 13.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 1.9 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 35.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 24.9 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 9.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 4.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 7.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 1.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 1.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.4 | 12.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 1.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 8.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 5.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 8.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 35.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 9.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 2.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.8 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 1.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 9.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |