Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
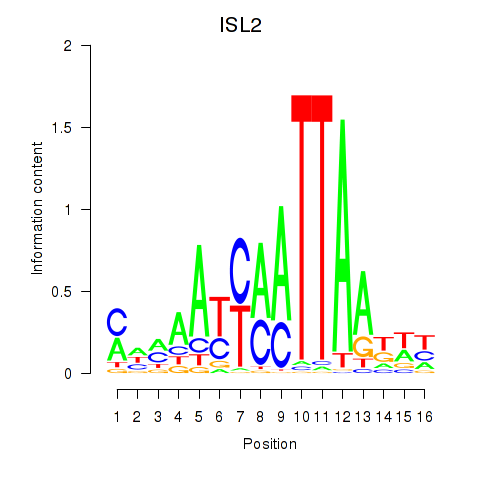
Results for ISL2
Z-value: 0.82
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.5 | ISL LIM homeobox 2 |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_95242044 | 13.50 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr10_-_95241951 | 13.06 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr3_+_172468505 | 12.61 |
ENST00000427830.1
ENST00000417960.1 ENST00000428567.1 ENST00000366090.2 ENST00000426894.1 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr2_-_10587897 | 12.36 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr3_+_172468472 | 11.58 |
ENST00000232458.5
ENST00000392692.3 |
ECT2
|
epithelial cell transforming sequence 2 oncogene |
| chr4_+_113568207 | 11.48 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr11_-_107729887 | 9.78 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr8_-_121457608 | 8.52 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr3_-_141747950 | 8.48 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_+_99127225 | 7.59 |
ENST00000370189.5
ENST00000529992.1 |
SNX7
|
sorting nexin 7 |
| chr12_+_64798095 | 7.34 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr7_-_16844611 | 7.30 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr11_+_101983176 | 7.25 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr12_+_104337515 | 7.20 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr3_+_139063372 | 6.09 |
ENST00000478464.1
|
MRPS22
|
mitochondrial ribosomal protein S22 |
| chr2_-_136633940 | 5.88 |
ENST00000264156.2
|
MCM6
|
minichromosome maintenance complex component 6 |
| chr2_+_86669118 | 5.69 |
ENST00000427678.1
ENST00000542128.1 |
KDM3A
|
lysine (K)-specific demethylase 3A |
| chr11_-_62609281 | 5.62 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr2_+_231921574 | 5.18 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr11_-_28129656 | 4.84 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr3_+_136649311 | 4.53 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr17_+_7155819 | 4.26 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr4_+_169418195 | 4.19 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_-_64225508 | 4.04 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_169013666 | 4.04 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr2_+_234668894 | 3.97 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr5_-_130500922 | 3.96 |
ENST00000513012.1
ENST00000508488.1 ENST00000506908.1 ENST00000304043.5 |
HINT1
|
histidine triad nucleotide binding protein 1 |
| chr11_-_327537 | 3.86 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr21_-_30445886 | 3.76 |
ENST00000431234.1
ENST00000540844.1 ENST00000286788.4 |
CCT8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr3_+_138327417 | 3.58 |
ENST00000338446.4
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_-_23058063 | 3.55 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr1_-_95391315 | 3.21 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr11_+_73498898 | 3.17 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr15_+_64680003 | 3.16 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr22_-_29107919 | 3.10 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr17_-_48785216 | 3.03 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr15_+_79166065 | 3.02 |
ENST00000559690.1
ENST00000559158.1 |
MORF4L1
|
mortality factor 4 like 1 |
| chr3_+_138327542 | 3.02 |
ENST00000360570.3
ENST00000393035.2 |
FAIM
|
Fas apoptotic inhibitory molecule |
| chr14_-_67878917 | 3.00 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr11_-_63376013 | 2.77 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr10_-_5046042 | 2.48 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr6_+_151561085 | 2.42 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr6_-_111804905 | 2.39 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_-_100872911 | 2.31 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr14_-_51027838 | 2.22 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr20_-_33735070 | 2.21 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_+_12693336 | 2.09 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr10_+_35484793 | 2.03 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chrX_-_77225135 | 1.87 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr7_-_45151272 | 1.80 |
ENST00000461363.1
ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4
|
transforming growth factor beta regulator 4 |
| chrX_+_102840408 | 1.77 |
ENST00000468024.1
ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4
|
transcription elongation factor A (SII)-like 4 |
| chr4_+_128702969 | 1.64 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr5_+_32788945 | 1.53 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chrX_+_108779004 | 1.37 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr12_+_26164645 | 1.37 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr17_-_10372875 | 1.35 |
ENST00000255381.2
|
MYH4
|
myosin, heavy chain 4, skeletal muscle |
| chr10_-_29923893 | 1.28 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr1_-_21377383 | 1.26 |
ENST00000374935.3
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr8_+_70404996 | 1.17 |
ENST00000402687.4
ENST00000419716.3 |
SULF1
|
sulfatase 1 |
| chr18_-_64271363 | 1.17 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr2_+_234580525 | 1.11 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_-_7307358 | 1.07 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr5_-_139726181 | 1.01 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr4_+_169418255 | 0.99 |
ENST00000505667.1
ENST00000511948.1 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_93257951 | 0.94 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr1_+_149871171 | 0.90 |
ENST00000369150.1
|
BOLA1
|
bolA family member 1 |
| chr11_-_13517565 | 0.90 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr4_+_88532028 | 0.89 |
ENST00000282478.7
|
DSPP
|
dentin sialophosphoprotein |
| chr17_-_3496171 | 0.89 |
ENST00000399756.4
|
TRPV1
|
transient receptor potential cation channel, subfamily V, member 1 |
| chrX_-_84634737 | 0.88 |
ENST00000262753.4
|
POF1B
|
premature ovarian failure, 1B |
| chr1_-_174992544 | 0.81 |
ENST00000476371.1
|
MRPS14
|
mitochondrial ribosomal protein S14 |
| chr2_+_234580499 | 0.81 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr12_+_75784850 | 0.77 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr2_-_163008903 | 0.77 |
ENST00000418842.2
ENST00000375497.3 |
GCG
|
glucagon |
| chr12_-_54689532 | 0.72 |
ENST00000540264.2
ENST00000312156.4 |
NFE2
|
nuclear factor, erythroid 2 |
| chr6_-_155776966 | 0.61 |
ENST00000159060.2
|
NOX3
|
NADPH oxidase 3 |
| chr1_-_202897724 | 0.60 |
ENST00000435533.3
ENST00000367258.1 |
KLHL12
|
kelch-like family member 12 |
| chr1_-_190446759 | 0.57 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr6_+_64345698 | 0.49 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr3_-_183145873 | 0.28 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr4_-_122085469 | 0.22 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr14_+_102276132 | 0.22 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr2_-_217560248 | 0.22 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr17_-_10325261 | 0.20 |
ENST00000403437.2
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr1_+_13421176 | 0.15 |
ENST00000376152.1
|
PRAMEF9
|
PRAME family member 9 |
| chr9_+_111624577 | 0.14 |
ENST00000333999.3
|
ACTL7A
|
actin-like 7A |
| chr19_+_4402659 | 0.12 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr11_+_113779704 | 0.12 |
ENST00000537778.1
|
HTR3B
|
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr2_+_234826016 | 0.11 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr9_-_95166841 | 0.10 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr7_+_141811539 | 0.09 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr1_+_171227069 | 0.09 |
ENST00000354841.4
|
FMO1
|
flavin containing monooxygenase 1 |
| chr12_-_51422017 | 0.07 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr14_+_38677123 | 0.03 |
ENST00000267377.2
|
SSTR1
|
somatostatin receptor 1 |
| chr1_+_149871135 | 0.02 |
ENST00000369152.5
|
BOLA1
|
bolA family member 1 |
| chr19_-_46234119 | 0.01 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.4 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 2.2 | 26.6 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 1.9 | 5.7 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 1.6 | 24.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 1.5 | 4.5 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 1.2 | 7.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.0 | 3.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 1.0 | 5.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.8 | 7.3 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.7 | 2.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.6 | 5.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 1.9 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) positive regulation of sperm motility(GO:1902093) |
| 0.5 | 7.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 2.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 2.5 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.3 | 4.8 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.3 | 4.0 | GO:0034392 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.3 | 0.9 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 0.9 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.3 | 3.8 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.3 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.3 | 0.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.3 | 1.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.3 | 18.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 5.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 2.0 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.2 | 8.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.8 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 2.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 7.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 3.9 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 3.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 3.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 3.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 4.0 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.1 | 5.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 3.6 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.1 | 5.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 1.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.4 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.9 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 5.8 | GO:0016050 | vesicle organization(GO:0016050) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 1.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 24.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 1.5 | 7.3 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.6 | 12.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 4.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 5.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 4.8 | GO:1990023 | kinetochore microtubule(GO:0005828) mitotic spindle midzone(GO:1990023) |
| 0.3 | 1.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 3.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 11.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 3.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 6.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.3 | 5.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.3 | 4.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 26.6 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 4.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 5.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 2.4 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 3.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 2.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 3.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 3.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 4.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 7.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.6 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 3.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 2.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 3.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 4.3 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 2.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 1.1 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.8 | 4.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.7 | 7.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.6 | 2.8 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.5 | 1.9 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 2.5 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.4 | 3.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 7.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 5.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.3 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 24.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 4.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 7.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 5.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.9 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.2 | 12.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 5.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 7.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 2.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 2.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 3.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 4.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 4.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 15.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 25.3 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.1 | 1.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 3.0 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 8.0 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 24.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 31.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 8.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 7.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 12.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.1 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 8.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.5 | 12.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 24.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 5.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 7.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 5.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 4.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 4.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 2.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 3.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 5.2 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 3.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 2.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |