Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
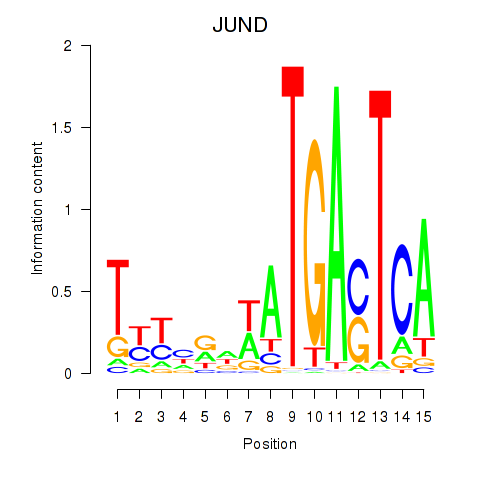
Results for JUND
Z-value: 0.40
Transcription factors associated with JUND
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
JUND
|
ENSG00000130522.4 | JunD proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| JUND | hg19_v2_chr19_-_18391708_18391739 | 0.60 | 8.5e-23 | Click! |
Activity profile of JUND motif
Sorted Z-values of JUND motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_84035905 | 21.77 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr4_-_84035868 | 21.00 |
ENST00000426923.2
ENST00000509973.1 |
PLAC8
|
placenta-specific 8 |
| chr3_-_18466026 | 17.64 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr5_-_140013275 | 13.87 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr22_-_37545972 | 13.70 |
ENST00000216223.5
|
IL2RB
|
interleukin 2 receptor, beta |
| chr15_+_89182178 | 12.32 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr16_+_23847339 | 12.15 |
ENST00000303531.7
|
PRKCB
|
protein kinase C, beta |
| chr15_+_89181974 | 11.99 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_+_54891495 | 10.44 |
ENST00000293373.6
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr1_-_153348067 | 9.83 |
ENST00000368737.3
|
S100A12
|
S100 calcium binding protein A12 |
| chr1_+_117297007 | 8.65 |
ENST00000369478.3
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr19_+_13842559 | 8.55 |
ENST00000586600.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr6_-_24936170 | 8.41 |
ENST00000538035.1
|
FAM65B
|
family with sequence similarity 65, member B |
| chr15_+_89182156 | 8.10 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr1_+_26605618 | 7.62 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr11_-_5255696 | 7.55 |
ENST00000292901.3
ENST00000417377.1 |
HBD
|
hemoglobin, delta |
| chr16_+_31366536 | 7.14 |
ENST00000562522.1
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr7_+_79998864 | 7.04 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr21_+_42733870 | 6.73 |
ENST00000330714.3
ENST00000436410.1 ENST00000435611.1 |
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr3_+_46412345 | 6.69 |
ENST00000292303.4
|
CCR5
|
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr11_+_67250490 | 6.59 |
ENST00000528641.2
ENST00000279146.3 |
AIP
|
aryl hydrocarbon receptor interacting protein |
| chr16_+_31366455 | 6.10 |
ENST00000268296.4
|
ITGAX
|
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr14_-_25103388 | 5.91 |
ENST00000526004.1
ENST00000415355.3 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr14_-_25103472 | 5.83 |
ENST00000216341.4
ENST00000382542.1 ENST00000382540.1 |
GZMB
|
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr14_+_58765103 | 5.80 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr14_-_23426322 | 5.52 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr14_-_23426270 | 5.42 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chrY_+_15016725 | 5.31 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr12_+_62654119 | 5.28 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr14_-_23426337 | 5.27 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr20_+_30946106 | 4.99 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr3_-_49395705 | 4.95 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr18_-_74207146 | 4.81 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr12_+_62654155 | 4.66 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr3_-_105588231 | 4.52 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr22_+_23101182 | 4.47 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr22_+_22707260 | 4.44 |
ENST00000390293.1
|
IGLV5-48
|
immunoglobulin lambda variable 5-48 (non-functional) |
| chr12_-_8218997 | 4.06 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr16_+_28996114 | 3.90 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr5_-_141061759 | 3.43 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr10_+_17270214 | 3.27 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr5_-_141061777 | 3.26 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr3_-_52713729 | 3.12 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr14_-_24711470 | 3.11 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr5_-_138533973 | 3.06 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr3_+_30647994 | 3.03 |
ENST00000295754.5
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa) |
| chr8_+_128426535 | 2.90 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr2_+_201980827 | 2.82 |
ENST00000309955.3
ENST00000443227.1 ENST00000341222.6 ENST00000355558.4 ENST00000340870.5 ENST00000341582.6 |
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr9_-_73029540 | 2.68 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr3_-_122512619 | 2.62 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr19_-_54872556 | 2.55 |
ENST00000444687.1
|
LAIR1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chr1_-_205744574 | 2.53 |
ENST00000367139.3
ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr1_-_6662919 | 2.51 |
ENST00000377658.4
ENST00000377663.3 |
KLHL21
|
kelch-like family member 21 |
| chr10_-_81708854 | 2.50 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr16_-_67281413 | 2.47 |
ENST00000258201.4
|
FHOD1
|
formin homology 2 domain containing 1 |
| chr3_-_150996239 | 2.46 |
ENST00000309170.3
|
P2RY14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr11_-_2162162 | 2.40 |
ENST00000381389.1
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr12_-_8765446 | 2.40 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chrX_-_154563889 | 2.39 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr17_+_1936687 | 2.39 |
ENST00000570477.1
|
DPH1
|
diphthamide biosynthesis 1 |
| chr12_+_9142131 | 2.34 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr5_-_138534071 | 2.22 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chrX_-_51797351 | 2.11 |
ENST00000375644.3
|
RP11-114H20.1
|
RP11-114H20.1 |
| chr14_+_73525144 | 2.07 |
ENST00000261973.7
ENST00000540173.1 |
RBM25
|
RNA binding motif protein 25 |
| chr2_+_169926047 | 1.92 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr18_-_19283649 | 1.88 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr12_+_52695617 | 1.84 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr1_+_41204506 | 1.81 |
ENST00000525290.1
ENST00000530965.1 ENST00000416859.2 ENST00000308733.5 |
NFYC
|
nuclear transcription factor Y, gamma |
| chrX_+_51942963 | 1.65 |
ENST00000375625.3
|
RP11-363G10.2
|
RP11-363G10.2 |
| chr14_+_73525229 | 1.57 |
ENST00000527432.1
ENST00000531500.1 ENST00000525321.1 ENST00000526754.1 |
RBM25
|
RNA binding motif protein 25 |
| chr4_-_100356291 | 1.55 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_+_149539767 | 1.48 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr1_+_160709029 | 1.45 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr3_+_127771212 | 1.35 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr2_+_102615416 | 1.27 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr20_+_48429233 | 1.26 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chrX_-_153718953 | 1.25 |
ENST00000369649.4
ENST00000393586.1 |
SLC10A3
|
solute carrier family 10, member 3 |
| chr19_+_11546093 | 1.14 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr15_-_42565606 | 1.12 |
ENST00000307216.6
ENST00000448392.1 |
TMEM87A
|
transmembrane protein 87A |
| chr20_+_48429356 | 1.09 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr19_+_35820064 | 0.98 |
ENST00000341773.6
ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22
|
CD22 molecule |
| chr5_-_135701164 | 0.96 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr8_-_17752912 | 0.95 |
ENST00000398054.1
ENST00000381840.2 |
FGL1
|
fibrinogen-like 1 |
| chrX_+_78200829 | 0.93 |
ENST00000544091.1
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr4_-_100356551 | 0.92 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr16_+_28996364 | 0.86 |
ENST00000564277.1
|
LAT
|
linker for activation of T cells |
| chr3_+_45067659 | 0.73 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr12_-_8380183 | 0.59 |
ENST00000442295.2
ENST00000307435.6 ENST00000538603.1 |
FAM90A1
|
family with sequence similarity 90, member A1 |
| chr6_-_44281043 | 0.59 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr10_-_14996070 | 0.58 |
ENST00000378258.1
ENST00000453695.2 ENST00000378246.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr2_+_161993412 | 0.56 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr10_-_14996017 | 0.55 |
ENST00000378241.1
ENST00000456122.1 ENST00000418843.1 ENST00000378249.1 ENST00000396817.2 ENST00000378255.1 ENST00000378254.1 ENST00000378278.2 ENST00000357717.2 |
DCLRE1C
|
DNA cross-link repair 1C |
| chr1_+_160709055 | 0.54 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_223889310 | 0.50 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr1_+_207262881 | 0.47 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr8_-_17752996 | 0.47 |
ENST00000381841.2
ENST00000427924.1 |
FGL1
|
fibrinogen-like 1 |
| chr22_-_39715600 | 0.47 |
ENST00000427905.1
ENST00000402527.1 ENST00000216146.4 |
RPL3
|
ribosomal protein L3 |
| chr10_-_74114714 | 0.43 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr18_+_32290218 | 0.42 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr17_-_7590745 | 0.40 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr17_-_45899126 | 0.38 |
ENST00000007414.3
ENST00000392507.3 |
OSBPL7
|
oxysterol binding protein-like 7 |
| chr14_+_73525265 | 0.32 |
ENST00000525161.1
|
RBM25
|
RNA binding motif protein 25 |
| chr11_-_2162468 | 0.29 |
ENST00000434045.2
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr17_+_30771279 | 0.28 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr1_-_1342617 | 0.24 |
ENST00000482352.1
ENST00000344843.7 |
MRPL20
|
mitochondrial ribosomal protein L20 |
| chr15_+_75335604 | 0.18 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr3_+_155860751 | 0.13 |
ENST00000471742.1
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr2_+_113670548 | 0.10 |
ENST00000263326.3
ENST00000352179.3 ENST00000349806.3 ENST00000353225.3 |
IL37
|
interleukin 37 |
| chr17_+_41052808 | 0.07 |
ENST00000592383.1
ENST00000253801.2 ENST00000585489.1 |
G6PC
|
glucose-6-phosphatase, catalytic subunit |
| chr17_-_59668550 | 0.07 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr20_-_17539456 | 0.05 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr3_+_11178779 | 0.03 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr12_-_7818474 | 0.02 |
ENST00000229304.4
|
APOBEC1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr9_-_95432536 | 0.00 |
ENST00000287996.3
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of JUND
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 32.4 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 4.0 | 12.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 3.5 | 13.9 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 2.6 | 10.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) actin polymerization-dependent cell motility(GO:0070358) |
| 2.5 | 9.9 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 2.4 | 42.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 2.0 | 13.7 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 1.7 | 11.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.7 | 5.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 1.5 | 5.8 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 1.4 | 8.6 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 1.4 | 16.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 1.2 | 7.0 | GO:0071724 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 1.1 | 6.6 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 1.0 | 3.0 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.0 | 5.0 | GO:0009608 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 1.0 | 6.7 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 1.0 | 6.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.8 | 2.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.6 | 2.5 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.6 | 6.7 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.5 | 3.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.5 | 7.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 2.7 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.4 | 4.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.3 | 2.4 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.3 | 1.3 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.3 | 2.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 2.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 3.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 2.8 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.3 | 4.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.3 | 1.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 2.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 9.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 3.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.4 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.9 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 2.4 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 13.2 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) |
| 0.1 | 2.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 4.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 16.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 7.6 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.1 | 0.7 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.1 | 2.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 5.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.5 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 4.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 4.8 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 1.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.9 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.5 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 2.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.8 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.4 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 3.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 4.5 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 3.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 5.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 13.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.8 | 16.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.6 | 6.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.7 | 10.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.6 | 3.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.5 | 7.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.5 | 5.0 | GO:0097413 | Lewy body(GO:0097413) |
| 0.5 | 32.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.4 | 17.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.4 | 42.8 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.3 | 3.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 16.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 2.5 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 13.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.3 | 8.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.3 | 7.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.7 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 2.8 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 2.5 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 2.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 1.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 5.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.1 | 3.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 5.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 6.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 17.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 9.3 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 5.3 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 6.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 3.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 32.4 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 4.6 | 13.9 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 4.0 | 12.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 3.4 | 13.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 1.7 | 9.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.2 | 7.0 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.8 | 3.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.6 | 2.5 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.6 | 7.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 3.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 7.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.5 | 6.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.5 | 9.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.5 | 6.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.4 | 2.5 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.4 | 1.9 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.4 | 5.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 2.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 1.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 4.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.3 | 1.9 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.3 | 6.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 1.3 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 7.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.2 | 2.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 1.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 13.1 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 5.8 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 2.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 4.8 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 5.3 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 52.5 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 4.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 3.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 15.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 5.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 3.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 5.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 4.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 14.4 | GO:0004872 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 16.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.4 | 13.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.4 | 11.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.3 | 13.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 12.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 14.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 6.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 5.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 2.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 8.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 1.2 | 13.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.5 | 5.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 39.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.4 | 12.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.4 | 9.7 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.3 | 2.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 12.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 12.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 21.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 14.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 3.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 4.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 1.5 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.2 | 3.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 8.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 1.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 7.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 6.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |