Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
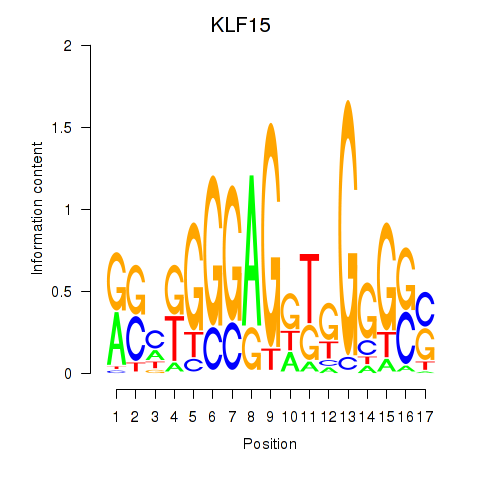
Results for KLF15
Z-value: 0.30
Transcription factors associated with KLF15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF15
|
ENSG00000163884.3 | Kruppel like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF15 | hg19_v2_chr3_-_126076264_126076305 | 0.50 | 4.3e-15 | Click! |
Activity profile of KLF15 motif
Sorted Z-values of KLF15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_77681075 | 17.83 |
ENST00000397549.2
|
CTD-2116F7.1
|
CTD-2116F7.1 |
| chr6_+_111580508 | 12.53 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr12_+_53773944 | 11.63 |
ENST00000551969.1
ENST00000327443.4 |
SP1
|
Sp1 transcription factor |
| chrX_+_16737718 | 10.95 |
ENST00000380155.3
|
SYAP1
|
synapse associated protein 1 |
| chr22_+_29469100 | 10.05 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr5_+_17217669 | 9.09 |
ENST00000322611.3
|
BASP1
|
brain abundant, membrane attached signal protein 1 |
| chrX_+_23352133 | 8.58 |
ENST00000379361.4
|
PTCHD1
|
patched domain containing 1 |
| chr6_+_43543864 | 8.50 |
ENST00000372236.4
ENST00000535400.1 |
POLH
|
polymerase (DNA directed), eta |
| chr11_-_64410787 | 8.27 |
ENST00000301894.2
|
NRXN2
|
neurexin 2 |
| chr5_-_11904100 | 8.08 |
ENST00000359640.2
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr15_-_45480153 | 8.01 |
ENST00000560471.1
ENST00000560540.1 |
SHF
|
Src homology 2 domain containing F |
| chr10_+_74451883 | 7.71 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr7_+_65338230 | 7.50 |
ENST00000360768.3
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr2_-_220173685 | 7.44 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr7_-_143059780 | 7.38 |
ENST00000409578.1
ENST00000409346.1 |
FAM131B
|
family with sequence similarity 131, member B |
| chr5_-_11904152 | 7.14 |
ENST00000304623.8
ENST00000458100.2 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr19_-_55866104 | 7.12 |
ENST00000326529.4
|
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr19_-_47975417 | 7.10 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr3_+_101546827 | 7.04 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr6_+_43543942 | 7.03 |
ENST00000372226.1
ENST00000443535.1 |
POLH
|
polymerase (DNA directed), eta |
| chr1_+_228353495 | 7.01 |
ENST00000366711.3
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr10_+_76871353 | 7.01 |
ENST00000542569.1
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr12_-_123215306 | 6.99 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr14_-_21493884 | 6.80 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr18_-_51750948 | 6.58 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr11_+_17756279 | 6.09 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr19_-_49576198 | 6.06 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr12_-_54982300 | 6.04 |
ENST00000547431.1
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr2_-_9143786 | 5.95 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chr16_+_56225248 | 5.90 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr10_+_76871454 | 5.79 |
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr8_+_9413410 | 5.60 |
ENST00000520408.1
ENST00000310430.6 ENST00000522110.1 |
TNKS
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr12_+_53443680 | 5.54 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr19_-_38806560 | 5.52 |
ENST00000591755.1
ENST00000337679.8 ENST00000339413.6 |
YIF1B
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr14_-_21493649 | 5.50 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr2_-_176866978 | 5.49 |
ENST00000392540.2
ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715
|
KIAA1715 |
| chr12_+_9067123 | 5.35 |
ENST00000543824.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr12_+_53443963 | 5.31 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_+_14989076 | 5.25 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr19_+_10531150 | 5.18 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr1_-_32229934 | 5.15 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr16_+_50187556 | 5.05 |
ENST00000561678.1
ENST00000357464.3 |
PAPD5
|
PAP associated domain containing 5 |
| chr11_-_62457371 | 4.85 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr12_+_9067327 | 4.83 |
ENST00000433083.2
ENST00000544916.1 ENST00000544539.1 ENST00000539063.1 |
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr18_+_56530136 | 4.79 |
ENST00000591083.1
|
ZNF532
|
zinc finger protein 532 |
| chr1_-_32229523 | 4.77 |
ENST00000398547.1
ENST00000373655.2 ENST00000373658.3 ENST00000257070.4 |
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr14_+_32546274 | 4.75 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr3_+_238273 | 4.70 |
ENST00000256509.2
|
CHL1
|
cell adhesion molecule L1-like |
| chr9_-_111929560 | 4.66 |
ENST00000561981.2
|
FRRS1L
|
ferric-chelate reductase 1-like |
| chr2_+_5832799 | 4.66 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr3_-_52001448 | 4.58 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr21_+_27107672 | 4.55 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr17_-_78450398 | 4.53 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr12_-_54982420 | 4.49 |
ENST00000257905.8
|
PPP1R1A
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr3_-_149688502 | 4.49 |
ENST00000481767.1
ENST00000475518.1 |
PFN2
|
profilin 2 |
| chrX_-_153881842 | 4.45 |
ENST00000369585.3
ENST00000247306.4 |
CTAG2
|
cancer/testis antigen 2 |
| chr7_-_105029329 | 4.42 |
ENST00000393651.3
ENST00000460391.1 |
SRPK2
|
SRSF protein kinase 2 |
| chr4_+_55524085 | 4.40 |
ENST00000412167.2
ENST00000288135.5 |
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| chr19_-_46272462 | 4.38 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr14_-_21493123 | 4.37 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr19_-_55866061 | 4.36 |
ENST00000588572.2
ENST00000593184.1 ENST00000589467.1 |
COX6B2
|
cytochrome c oxidase subunit VIb polypeptide 2 (testis) |
| chr15_+_64752927 | 4.31 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr2_-_37193606 | 4.25 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr17_-_41984835 | 4.25 |
ENST00000520406.1
ENST00000518478.1 ENST00000522172.1 ENST00000461854.1 ENST00000521178.1 ENST00000520305.1 ENST00000523501.1 ENST00000520241.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr12_-_6798616 | 4.24 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_123849374 | 4.22 |
ENST00000602398.1
ENST00000602750.1 |
SBNO1
|
strawberry notch homolog 1 (Drosophila) |
| chr3_+_111718173 | 4.17 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr7_+_29519662 | 4.07 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr22_+_19118321 | 4.03 |
ENST00000399635.2
|
TSSK2
|
testis-specific serine kinase 2 |
| chr5_-_73937244 | 3.99 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr12_-_6798410 | 3.92 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr11_-_72353451 | 3.88 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr1_+_97187318 | 3.86 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr17_-_15168624 | 3.82 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chrX_+_70316005 | 3.81 |
ENST00000374259.3
|
FOXO4
|
forkhead box O4 |
| chr12_-_6798523 | 3.79 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr5_+_140254884 | 3.78 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr12_-_112819896 | 3.77 |
ENST00000377560.5
ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr3_+_46923670 | 3.71 |
ENST00000427125.2
ENST00000430002.2 |
PTH1R
|
parathyroid hormone 1 receptor |
| chr10_+_60272814 | 3.71 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr17_-_41985096 | 3.67 |
ENST00000269095.4
ENST00000523220.1 |
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr11_-_31839488 | 3.65 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chr11_-_8954491 | 3.65 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr3_+_9851632 | 3.53 |
ENST00000426895.4
|
TTLL3
|
tubulin tyrosine ligase-like family, member 3 |
| chr7_-_92157760 | 3.52 |
ENST00000248633.4
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr1_-_177133818 | 3.50 |
ENST00000424564.2
ENST00000361833.2 |
ASTN1
|
astrotactin 1 |
| chr19_+_50706866 | 3.48 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr1_-_241520525 | 3.47 |
ENST00000366565.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr6_+_31865552 | 3.43 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr18_-_21977748 | 3.42 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr14_+_24837226 | 3.33 |
ENST00000554050.1
ENST00000554903.1 ENST00000554779.1 ENST00000250373.4 ENST00000553708.1 |
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr1_-_47407111 | 3.29 |
ENST00000371904.4
|
CYP4A11
|
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr12_-_48213568 | 3.27 |
ENST00000080059.7
ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7
|
histone deacetylase 7 |
| chr18_-_57364588 | 3.27 |
ENST00000439986.4
|
CCBE1
|
collagen and calcium binding EGF domains 1 |
| chr15_-_34629922 | 3.23 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr17_-_4464081 | 3.23 |
ENST00000574154.1
|
GGT6
|
gamma-glutamyltransferase 6 |
| chr15_-_34630234 | 3.19 |
ENST00000558667.1
ENST00000561120.1 ENST00000559236.1 ENST00000397702.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr9_-_14313641 | 3.16 |
ENST00000380953.1
|
NFIB
|
nuclear factor I/B |
| chrX_+_51927919 | 3.13 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr17_-_4463856 | 3.12 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr7_+_94285637 | 3.11 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chrX_-_65858865 | 3.11 |
ENST00000374719.3
ENST00000450752.1 ENST00000451436.2 |
EDA2R
|
ectodysplasin A2 receptor |
| chr1_-_205313304 | 3.07 |
ENST00000539253.1
ENST00000607826.1 |
KLHDC8A
|
kelch domain containing 8A |
| chr7_-_767249 | 3.07 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr11_+_119076745 | 3.07 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr3_-_156272924 | 3.05 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr10_+_35416223 | 3.03 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr2_-_242254595 | 3.03 |
ENST00000441124.1
ENST00000391976.2 |
HDLBP
|
high density lipoprotein binding protein |
| chr20_-_45280091 | 3.03 |
ENST00000396360.1
ENST00000435032.1 ENST00000413164.2 ENST00000372121.1 ENST00000339636.3 |
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr20_-_45280066 | 3.02 |
ENST00000279027.4
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3 |
| chr1_-_151689259 | 2.95 |
ENST00000420342.1
ENST00000290583.4 |
CELF3
|
CUGBP, Elav-like family member 3 |
| chr17_+_36508111 | 2.94 |
ENST00000331159.5
ENST00000577233.1 |
SOCS7
|
suppressor of cytokine signaling 7 |
| chrX_-_48693955 | 2.88 |
ENST00000218230.5
|
PCSK1N
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr14_+_32546485 | 2.87 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr20_+_62711482 | 2.83 |
ENST00000336866.2
ENST00000355631.4 |
OPRL1
|
opiate receptor-like 1 |
| chrX_-_65859096 | 2.83 |
ENST00000456230.2
|
EDA2R
|
ectodysplasin A2 receptor |
| chrY_+_4868267 | 2.79 |
ENST00000333703.4
|
PCDH11Y
|
protocadherin 11 Y-linked |
| chr2_-_220174166 | 2.78 |
ENST00000409251.3
ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chr7_-_92157747 | 2.77 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr19_-_40791211 | 2.75 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr11_-_65640325 | 2.74 |
ENST00000307998.6
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr19_-_14117074 | 2.73 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr11_-_73309228 | 2.73 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr14_+_33408449 | 2.73 |
ENST00000346562.2
ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr1_-_241520385 | 2.70 |
ENST00000366564.1
|
RGS7
|
regulator of G-protein signaling 7 |
| chr6_+_41606176 | 2.64 |
ENST00000441667.1
ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI
|
MyoD family inhibitor |
| chr1_-_243418621 | 2.64 |
ENST00000366544.1
ENST00000366543.1 |
CEP170
|
centrosomal protein 170kDa |
| chr10_-_79397391 | 2.63 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr6_-_29595779 | 2.63 |
ENST00000355973.3
ENST00000377012.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_+_29519486 | 2.63 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr4_-_5890145 | 2.62 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr2_+_135676381 | 2.60 |
ENST00000537343.1
ENST00000295238.6 ENST00000264157.5 |
CCNT2
|
cyclin T2 |
| chr2_+_48757278 | 2.59 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr10_+_74927875 | 2.56 |
ENST00000242505.6
|
FAM149B1
|
family with sequence similarity 149, member B1 |
| chr22_+_29469012 | 2.56 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr3_+_12838161 | 2.55 |
ENST00000456430.2
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr2_-_25564750 | 2.55 |
ENST00000321117.5
|
DNMT3A
|
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr14_+_59655369 | 2.53 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr15_-_86338134 | 2.52 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr3_-_105587879 | 2.50 |
ENST00000264122.4
ENST00000403724.1 ENST00000405772.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr21_-_45681765 | 2.47 |
ENST00000431166.1
|
DNMT3L
|
DNA (cytosine-5-)-methyltransferase 3-like |
| chr8_+_30241995 | 2.46 |
ENST00000397323.4
ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr1_-_204654481 | 2.45 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr2_-_31361543 | 2.45 |
ENST00000349752.5
|
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr1_-_32801825 | 2.45 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr22_+_31090793 | 2.43 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr4_-_170947485 | 2.42 |
ENST00000504999.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr19_-_51456198 | 2.41 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5 |
| chr3_+_38495333 | 2.40 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr3_+_184279566 | 2.35 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chr8_+_55370487 | 2.34 |
ENST00000297316.4
|
SOX17
|
SRY (sex determining region Y)-box 17 |
| chr15_-_88799948 | 2.31 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr9_+_140772226 | 2.31 |
ENST00000277551.2
ENST00000371372.1 ENST00000277549.5 ENST00000371363.1 ENST00000371357.1 ENST00000371355.4 |
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr17_-_4545170 | 2.30 |
ENST00000576394.1
ENST00000574640.1 |
ALOX15
|
arachidonate 15-lipoxygenase |
| chr1_-_244013384 | 2.30 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr20_-_48099182 | 2.30 |
ENST00000371741.4
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr3_+_11034403 | 2.29 |
ENST00000287766.4
ENST00000425938.1 |
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter), member 1 |
| chrX_-_17878827 | 2.29 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr7_-_108096822 | 2.27 |
ENST00000379028.3
ENST00000413765.2 ENST00000379022.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr19_+_15218180 | 2.27 |
ENST00000342784.2
ENST00000597977.1 ENST00000600440.1 |
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr15_-_86338100 | 2.26 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr13_-_108867846 | 2.26 |
ENST00000442234.1
|
LIG4
|
ligase IV, DNA, ATP-dependent |
| chr7_-_108096765 | 2.25 |
ENST00000379024.4
ENST00000351718.4 |
NRCAM
|
neuronal cell adhesion molecule |
| chr9_+_101867387 | 2.25 |
ENST00000374990.2
ENST00000552516.1 |
TGFBR1
|
transforming growth factor, beta receptor 1 |
| chr3_-_188665428 | 2.22 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr12_+_57853918 | 2.22 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr1_-_113498943 | 2.20 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr11_-_117103208 | 2.17 |
ENST00000320934.3
ENST00000530269.1 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr11_-_6677018 | 2.16 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr9_-_84303269 | 2.16 |
ENST00000418319.1
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
| chr5_-_141257954 | 2.13 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr6_-_131384412 | 2.12 |
ENST00000445890.2
ENST00000368128.2 ENST00000337057.3 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_+_74702214 | 2.10 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr7_+_144052381 | 2.08 |
ENST00000498580.1
ENST00000056217.5 |
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr10_+_35415978 | 2.08 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_149068464 | 2.07 |
ENST00000367463.4
|
UST
|
uronyl-2-sulfotransferase |
| chr16_-_62070305 | 2.07 |
ENST00000584337.1
|
CDH8
|
cadherin 8, type 2 |
| chrX_+_11776278 | 2.06 |
ENST00000312196.4
ENST00000337339.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr1_+_154975110 | 2.06 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr14_+_24867992 | 2.05 |
ENST00000382554.3
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr2_+_24272543 | 2.04 |
ENST00000380991.4
|
FKBP1B
|
FK506 binding protein 1B, 12.6 kDa |
| chr3_-_10547333 | 2.03 |
ENST00000383800.4
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr3_-_49726486 | 2.01 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr1_+_41249539 | 2.01 |
ENST00000347132.5
ENST00000509682.2 |
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr10_-_133795416 | 2.00 |
ENST00000540159.1
ENST00000368636.4 |
BNIP3
|
BCL2/adenovirus E1B 19kDa interacting protein 3 |
| chr20_+_20348740 | 1.99 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr14_-_104313824 | 1.99 |
ENST00000553739.1
ENST00000202556.9 |
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B |
| chr19_+_45754505 | 1.98 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
| chr19_+_589893 | 1.98 |
ENST00000251287.2
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr22_+_39745930 | 1.96 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr1_-_179198702 | 1.95 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr4_-_39640700 | 1.93 |
ENST00000295958.5
|
SMIM14
|
small integral membrane protein 14 |
| chr21_-_40685504 | 1.93 |
ENST00000380800.3
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr6_+_44238203 | 1.92 |
ENST00000451188.2
|
TMEM151B
|
transmembrane protein 151B |
| chr18_+_13218769 | 1.91 |
ENST00000399848.3
ENST00000361205.4 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr7_+_66205643 | 1.90 |
ENST00000380828.2
ENST00000510829.2 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr7_-_94285402 | 1.89 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr1_-_227506158 | 1.88 |
ENST00000366769.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chrX_+_119495934 | 1.86 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr3_-_12200851 | 1.86 |
ENST00000287814.4
|
TIMP4
|
TIMP metallopeptidase inhibitor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 2.2 | 15.5 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 2.1 | 6.3 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 2.1 | 6.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 2.0 | 11.9 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 1.9 | 5.6 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.8 | 9.1 | GO:2001076 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 1.6 | 12.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 1.6 | 4.7 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 1.5 | 1.5 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 1.5 | 6.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.4 | 6.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 1.4 | 5.4 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 1.3 | 3.9 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 1.3 | 3.9 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.3 | 7.7 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 1.3 | 6.4 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 1.2 | 4.8 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 1.2 | 3.5 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 1.0 | 16.7 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 1.0 | 8.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 | 3.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) response to water-immersion restraint stress(GO:1990785) |
| 1.0 | 4.8 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.9 | 4.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.9 | 2.6 | GO:1904530 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.9 | 6.0 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 7.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.8 | 3.3 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.8 | 3.3 | GO:1905075 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.8 | 4.0 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.8 | 2.3 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) gall bladder development(GO:0061010) |
| 0.8 | 3.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.8 | 9.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.8 | 2.3 | GO:2001303 | cellular response to interleukin-13(GO:0035963) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.8 | 4.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.8 | 2.3 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.7 | 2.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.7 | 3.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.7 | 5.1 | GO:0043634 | polyadenylation-dependent ncRNA catabolic process(GO:0043634) |
| 0.7 | 2.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.7 | 4.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.7 | 2.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.7 | 2.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.7 | 3.3 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.6 | 4.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.6 | 2.6 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.6 | 2.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.6 | 7.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.6 | 2.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.6 | 2.3 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.6 | 8.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.6 | 9.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.6 | 2.2 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.5 | 2.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.5 | 2.1 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.5 | 2.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.5 | 3.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.5 | 2.0 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.5 | 1.9 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.5 | 3.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) positive regulation of axon regeneration(GO:0048680) action potential propagation(GO:0098870) |
| 0.5 | 5.9 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.5 | 6.3 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.5 | 4.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.4 | 4.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.4 | 1.7 | GO:0097368 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.4 | 3.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.4 | 2.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) cellular response to cobalt ion(GO:0071279) |
| 0.4 | 4.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 4.1 | GO:0034465 | response to carbon monoxide(GO:0034465) negative regulation of cell volume(GO:0045794) |
| 0.4 | 0.7 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.4 | 4.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 1.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.4 | 2.9 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.4 | 1.8 | GO:0072240 | thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 0.3 | 2.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.3 | 0.7 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.3 | 7.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 0.9 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.3 | 1.5 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.3 | 1.5 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.3 | 0.9 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.3 | 1.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 4.3 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.3 | 3.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 3.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.3 | 2.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 4.5 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.2 | 1.0 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 2.4 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.2 | 2.1 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.2 | 3.0 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 2.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 5.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.2 | 7.9 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.2 | 0.4 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.2 | 0.4 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.2 | 2.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.7 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.2 | 1.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 6.4 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.2 | 6.0 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.2 | 13.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 3.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 1.8 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.2 | 1.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 2.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.9 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 5.2 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.2 | 2.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.1 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.2 | 1.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 3.5 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 3.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.3 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 | 2.6 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.4 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 12.6 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 1.3 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.1 | 1.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 2.3 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 5.9 | GO:0043278 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 2.4 | GO:0098761 | interleukin-7-mediated signaling pathway(GO:0038111) response to interleukin-7(GO:0098760) cellular response to interleukin-7(GO:0098761) |
| 0.1 | 1.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 2.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 2.2 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 2.4 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 1.9 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 3.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 3.0 | GO:0048265 | response to pain(GO:0048265) |
| 0.1 | 0.6 | GO:0072017 | distal tubule development(GO:0072017) |
| 0.1 | 1.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 5.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 3.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.7 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.1 | 1.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.6 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 6.4 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.5 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 2.3 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.8 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 2.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 1.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 1.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 3.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 1.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.1 | 2.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.2 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 10.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 4.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.7 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 4.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.1 | 2.7 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 2.0 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.0 | 3.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 2.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.2 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 2.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 5.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.7 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 2.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.8 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.6 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 1.4 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 8.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 1.9 | GO:0042698 | ovulation cycle(GO:0042698) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 2.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.5 | GO:0048048 | embryonic eye morphogenesis(GO:0048048) |
| 0.0 | 1.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 1.7 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.5 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.0 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 2.0 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.9 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.4 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.0 | 1.8 | GO:0030100 | regulation of endocytosis(GO:0030100) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.7 | GO:1990246 | uniplex complex(GO:1990246) |
| 1.6 | 11.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.9 | 2.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.7 | 3.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.6 | 4.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.5 | 4.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.5 | 20.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.5 | 7.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.5 | 6.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.4 | 2.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.4 | 5.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.4 | 2.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.4 | 3.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.4 | 1.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.4 | 7.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 5.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 6.1 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.3 | 2.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 4.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 39.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 1.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 4.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 1.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 2.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 7.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.6 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.2 | 2.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 3.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 11.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 28.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 3.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 7.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 3.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 14.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 11.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 4.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 1.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 6.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 1.4 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 8.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 8.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 3.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 21.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 3.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 3.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 2.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 3.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 10.2 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 1.1 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 3.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 3.7 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.8 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 3.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.3 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 4.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.4 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.5 | GO:1902554 | serine/threonine protein kinase complex(GO:1902554) |
| 0.0 | 1.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.5 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 57.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 1.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 12.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.9 | 7.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 1.5 | 6.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.3 | 10.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.2 | 4.8 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.9 | 2.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 3.5 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.8 | 3.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.8 | 2.3 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.7 | 6.0 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.7 | 3.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.7 | 3.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.7 | 7.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.7 | 20.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.7 | 5.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.7 | 5.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.7 | 6.6 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.6 | 3.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.6 | 3.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.6 | 4.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.6 | 6.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.6 | 6.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 3.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 5.9 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 8.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.5 | 2.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 2.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.5 | 6.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.4 | 2.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 1.7 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.4 | 5.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.4 | 6.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 9.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.4 | 2.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.4 | 1.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.4 | 1.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 1.7 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.3 | 4.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 2.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 3.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 1.5 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.3 | 0.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.3 | 0.9 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.3 | 1.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.3 | 0.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 1.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.3 | 2.9 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.3 | 4.4 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 1.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.2 | 12.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 2.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.0 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 3.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 2.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 3.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 7.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 2.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 2.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 8.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 3.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 5.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 3.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 5.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 4.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 0.7 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
| 0.2 | 2.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 1.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 4.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 17.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 3.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 7.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 15.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 8.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 3.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.7 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 3.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 3.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 4.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 3.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 3.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 1.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 7.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 5.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 4.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 4.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.6 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 8.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 8.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.9 | GO:0015077 | monovalent inorganic cation transmembrane transporter activity(GO:0015077) |
| 0.0 | 1.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.5 | 3.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.4 | 4.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.4 | 3.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.3 | 8.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 12.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 4.8 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 7.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 10.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.2 | 5.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.2 | 3.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 19.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 9.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 4.9 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 9.8 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 3.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 2.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 8.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 10.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.4 | 11.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 6.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 12.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 6.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 3.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 6.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 6.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 5.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.2 | 1.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 3.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.8 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 3.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.0 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 4.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 17.3 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.1 | 3.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.7 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 6.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 2.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 3.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 2.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 5.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.1 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 11.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 4.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 2.5 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 6.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME PKB MEDIATED EVENTS | Genes involved in PKB-mediated events |