Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
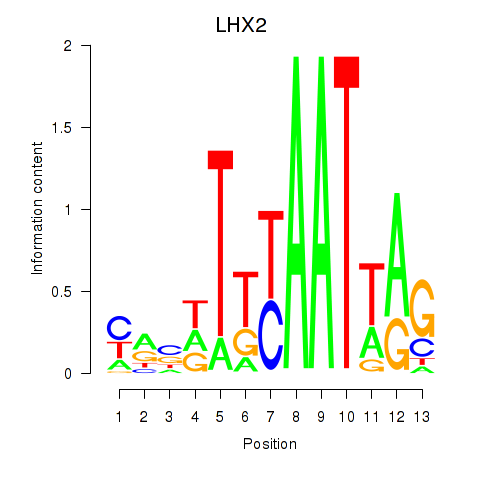
Results for LHX2
Z-value: 0.26
Transcription factors associated with LHX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
LHX2
|
ENSG00000106689.6 | LIM homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX2 | hg19_v2_chr9_+_126777676_126777700, hg19_v2_chr9_+_126773880_126773895 | -0.04 | 5.9e-01 | Click! |
Activity profile of LHX2 motif
Sorted Z-values of LHX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_6309517 | 12.86 |
ENST00000382519.4
ENST00000009180.4 |
CD9
|
CD9 molecule |
| chr7_-_5998714 | 4.98 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr6_+_125524785 | 4.24 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr21_-_43735628 | 4.08 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr4_+_70894130 | 3.57 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr21_-_43735446 | 3.54 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr14_+_56078695 | 3.52 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr10_-_116286656 | 3.44 |
ENST00000428430.1
ENST00000369266.3 ENST00000392952.3 |
ABLIM1
|
actin binding LIM protein 1 |
| chr1_-_28527152 | 3.31 |
ENST00000321830.5
|
AL353354.1
|
Uncharacterized protein |
| chr14_-_69864993 | 3.22 |
ENST00000555373.1
|
ERH
|
enhancer of rudimentary homolog (Drosophila) |
| chrX_+_19373700 | 3.18 |
ENST00000379804.1
|
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr4_+_70916119 | 3.10 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr3_-_149095652 | 3.00 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr17_+_48823975 | 2.97 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr6_+_30848557 | 2.94 |
ENST00000460944.2
ENST00000324771.8 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_153518270 | 2.91 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr8_-_109260897 | 2.75 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr16_-_66864806 | 2.48 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr3_-_131221790 | 2.32 |
ENST00000512877.1
ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr18_+_3247779 | 2.27 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr2_-_55237484 | 2.23 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr13_+_73632897 | 2.22 |
ENST00000377687.4
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr4_+_71226468 | 2.13 |
ENST00000226460.4
|
SMR3A
|
submaxillary gland androgen regulated protein 3A |
| chr2_+_90139056 | 2.07 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr4_+_128554081 | 2.04 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr1_-_147142557 | 2.03 |
ENST00000369238.6
|
ACP6
|
acid phosphatase 6, lysophosphatidic |
| chr14_-_106926724 | 2.00 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr17_+_17082842 | 2.00 |
ENST00000579361.1
|
MPRIP
|
myosin phosphatase Rho interacting protein |
| chr19_+_42212526 | 1.97 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr6_+_26199737 | 1.94 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr2_+_109204909 | 1.82 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr12_+_6554021 | 1.80 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr2_-_55496174 | 1.80 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr7_-_54826869 | 1.78 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr7_-_151217166 | 1.76 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr1_+_62439037 | 1.68 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr3_+_108541545 | 1.68 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr19_+_22235310 | 1.55 |
ENST00000600162.1
|
ZNF257
|
zinc finger protein 257 |
| chr7_-_54826920 | 1.48 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr1_+_152975488 | 1.46 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr3_+_108541608 | 1.40 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr11_+_64001962 | 1.38 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chrX_-_11284095 | 1.33 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr13_-_99910673 | 1.32 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr21_-_27423339 | 1.31 |
ENST00000415997.1
|
APP
|
amyloid beta (A4) precursor protein |
| chr2_-_55276320 | 1.23 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chrX_+_105937068 | 1.20 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr2_-_89157161 | 1.13 |
ENST00000390237.2
|
IGKC
|
immunoglobulin kappa constant |
| chr1_-_89591749 | 1.11 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr20_-_31989307 | 1.07 |
ENST00000473997.1
ENST00000544843.1 ENST00000346416.2 ENST00000357886.4 ENST00000339269.5 |
CDK5RAP1
|
CDK5 regulatory subunit associated protein 1 |
| chr19_+_42212501 | 1.06 |
ENST00000398599.4
|
CEACAM5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr2_+_234627424 | 1.01 |
ENST00000373409.3
|
UGT1A4
|
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr7_-_99716952 | 0.97 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrM_+_12331 | 0.92 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_+_180682720 | 0.89 |
ENST00000599439.1
|
AC008443.1
|
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr13_+_24844819 | 0.87 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr12_+_16064106 | 0.87 |
ENST00000428559.2
|
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr1_+_145883868 | 0.86 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr11_-_104827425 | 0.84 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_-_26199499 | 0.83 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr12_-_118797475 | 0.83 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr3_+_38017264 | 0.83 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr4_+_88532028 | 0.82 |
ENST00000282478.7
|
DSPP
|
dentin sialophosphoprotein |
| chr8_-_19459993 | 0.81 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr21_-_35014027 | 0.81 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr2_-_231084617 | 0.80 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr2_+_120436733 | 0.79 |
ENST00000401466.1
ENST00000424086.1 ENST00000272521.6 |
TMEM177
|
transmembrane protein 177 |
| chr2_+_103035102 | 0.77 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr4_+_147096837 | 0.76 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr18_+_29027696 | 0.76 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr7_-_22234381 | 0.74 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_+_112408768 | 0.73 |
ENST00000368656.2
ENST00000604268.1 |
FAM229B
|
family with sequence similarity 229, member B |
| chr7_-_99717463 | 0.69 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_160709055 | 0.65 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr3_+_35722487 | 0.64 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr13_-_99959641 | 0.60 |
ENST00000376414.4
|
GPR183
|
G protein-coupled receptor 183 |
| chr2_-_10587897 | 0.60 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr8_+_22424551 | 0.58 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr10_+_124320156 | 0.53 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr11_+_120973375 | 0.53 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr1_+_95616933 | 0.51 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr2_-_99279928 | 0.50 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr20_+_56964169 | 0.48 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr6_-_131291572 | 0.47 |
ENST00000529208.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_+_160709029 | 0.47 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chr7_+_101460882 | 0.44 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr15_-_72563585 | 0.43 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr11_-_104972158 | 0.43 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr10_+_95326416 | 0.41 |
ENST00000371481.4
ENST00000371483.4 ENST00000604414.1 |
FFAR4
|
free fatty acid receptor 4 |
| chr10_-_61900762 | 0.40 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_105416039 | 0.39 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr5_+_179921344 | 0.36 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr8_-_82395461 | 0.33 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr2_+_87135076 | 0.32 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr10_-_14372870 | 0.31 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr12_+_9980069 | 0.31 |
ENST00000354855.3
ENST00000324214.4 ENST00000279544.3 |
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1 |
| chr21_+_34619079 | 0.31 |
ENST00000433395.2
|
AP000295.9
|
AP000295.9 |
| chrX_+_52513455 | 0.30 |
ENST00000446098.1
|
XAGE1C
|
X antigen family, member 1C |
| chr17_-_38859996 | 0.30 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr8_+_76452097 | 0.28 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr2_-_175712270 | 0.28 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr4_-_153332886 | 0.28 |
ENST00000603841.1
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr20_+_43029911 | 0.26 |
ENST00000443598.2
ENST00000316099.4 ENST00000415691.2 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr2_-_231084659 | 0.25 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr10_-_29923893 | 0.24 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr18_+_34124507 | 0.24 |
ENST00000591635.1
|
FHOD3
|
formin homology 2 domain containing 3 |
| chr4_-_70626314 | 0.22 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr11_-_62323702 | 0.21 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr2_+_68961934 | 0.20 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 0.20 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr7_-_142232071 | 0.18 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr9_+_108320392 | 0.17 |
ENST00000602661.1
ENST00000223528.2 ENST00000448551.2 ENST00000540160.1 |
FKTN
|
fukutin |
| chr2_-_166930131 | 0.15 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr20_+_30697298 | 0.14 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr7_-_103848405 | 0.13 |
ENST00000447452.2
ENST00000545943.1 ENST00000297431.4 |
ORC5
|
origin recognition complex, subunit 5 |
| chr14_-_107211459 | 0.13 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chrX_-_19988382 | 0.12 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr1_+_52682052 | 0.12 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr12_+_64173583 | 0.10 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr6_-_26199471 | 0.09 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr9_+_116207007 | 0.09 |
ENST00000374140.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr16_+_24266874 | 0.08 |
ENST00000005284.3
|
CACNG3
|
calcium channel, voltage-dependent, gamma subunit 3 |
| chr4_+_69313145 | 0.05 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr1_+_160709076 | 0.05 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr16_-_23652570 | 0.04 |
ENST00000261584.4
|
PALB2
|
partner and localizer of BRCA2 |
| chr12_-_10955226 | 0.04 |
ENST00000240687.2
|
TAS2R7
|
taste receptor, type 2, member 7 |
| chr12_-_11548496 | 0.02 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr6_-_27841289 | 0.01 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
Network of associatons between targets according to the STRING database.
First level regulatory network of LHX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 12.9 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.5 | 2.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 3.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.4 | 3.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 1.8 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.4 | 3.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.3 | 2.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 2.8 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.3 | 1.3 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.2 | 2.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 1.8 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 1.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 0.8 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 0.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.2 | 1.8 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.2 | 1.0 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.2 | 6.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.2 | 1.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.1 | 0.6 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.9 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 2.0 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 1.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 3.0 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.4 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 3.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.4 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 3.2 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 1.8 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.5 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 2.0 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 4.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 3.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 3.3 | GO:0071806 | protein transmembrane transport(GO:0071806) |
| 0.0 | 6.8 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 2.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 4.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 2.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.8 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 2.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 2.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 2.9 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 1.7 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.2 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 3.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.9 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.3 | 3.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.3 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 3.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 3.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 3.0 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 3.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 2.8 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.7 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.8 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 3.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 2.3 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 3.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.9 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 6.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 10.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.6 | 2.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 3.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.3 | 3.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 0.9 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.3 | 0.8 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.2 | 1.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 2.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.2 | 3.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 2.9 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 2.0 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 3.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.7 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 12.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 3.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 1.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.1 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 9.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.0 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 12.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 7.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 4.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 3.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 15.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.8 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 3.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 4.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 3.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 2.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |