Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
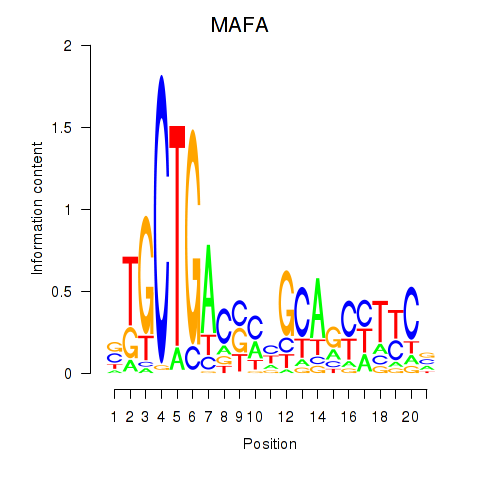
Results for MAFA
Z-value: 0.94
Transcription factors associated with MAFA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MAFA
|
ENSG00000182759.3 | MAF bZIP transcription factor A |
Activity profile of MAFA motif
Sorted Z-values of MAFA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_33182823 | 19.72 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr12_+_79258444 | 19.04 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr12_+_79258547 | 17.15 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr16_+_56225248 | 15.08 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr21_+_41239243 | 14.39 |
ENST00000328619.5
|
PCP4
|
Purkinje cell protein 4 |
| chr11_-_777467 | 13.54 |
ENST00000397472.2
ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1
|
Parkinson disease 7 domain containing 1 |
| chr12_-_105478339 | 13.45 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr10_+_124221036 | 13.06 |
ENST00000368984.3
|
HTRA1
|
HtrA serine peptidase 1 |
| chr15_+_80696666 | 12.96 |
ENST00000303329.4
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_163039143 | 12.90 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr9_-_100000957 | 11.93 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr10_-_79397391 | 11.50 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_+_141121164 | 11.49 |
ENST00000510338.1
ENST00000504673.1 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chrX_-_92928557 | 11.15 |
ENST00000373079.3
ENST00000475430.2 |
NAP1L3
|
nucleosome assembly protein 1-like 3 |
| chr12_-_123215306 | 10.76 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr13_+_53226963 | 10.12 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr6_-_74161977 | 10.10 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr16_-_46865047 | 9.81 |
ENST00000394806.2
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr3_+_196439170 | 9.40 |
ENST00000392391.3
ENST00000314118.4 |
PIGX
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr5_-_118324200 | 8.40 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr3_-_196439065 | 7.94 |
ENST00000399942.4
ENST00000409690.3 |
CEP19
|
centrosomal protein 19kDa |
| chr8_+_85095769 | 7.19 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr7_-_73133959 | 7.01 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr1_-_57045228 | 6.72 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr22_-_28197486 | 6.67 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chrX_-_47509887 | 6.62 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr2_+_191745535 | 6.47 |
ENST00000320717.3
|
GLS
|
glutaminase |
| chr5_-_114505624 | 6.46 |
ENST00000513154.1
|
TRIM36
|
tripartite motif containing 36 |
| chr10_+_120967072 | 6.43 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr8_+_85095497 | 6.21 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr17_+_41363854 | 6.10 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr15_-_66858298 | 6.07 |
ENST00000537670.1
|
LCTL
|
lactase-like |
| chr7_-_135412925 | 5.85 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr11_+_107461948 | 5.81 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr16_+_70258261 | 5.48 |
ENST00000594734.1
|
FKSG63
|
FKSG63 |
| chrX_-_107975917 | 5.46 |
ENST00000563887.1
|
RP6-24A23.6
|
Uncharacterized protein |
| chr2_+_118846008 | 5.39 |
ENST00000245787.4
|
INSIG2
|
insulin induced gene 2 |
| chr1_-_216978709 | 5.39 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr5_-_179285785 | 5.30 |
ENST00000520698.1
ENST00000518235.1 ENST00000376931.2 ENST00000518219.1 ENST00000521333.1 ENST00000523084.1 |
C5orf45
|
chromosome 5 open reading frame 45 |
| chrX_-_43741594 | 5.28 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr8_+_85095553 | 5.12 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr1_+_156124162 | 4.90 |
ENST00000368282.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr5_-_179285848 | 4.82 |
ENST00000403396.2
ENST00000292586.6 |
C5orf45
|
chromosome 5 open reading frame 45 |
| chr1_+_66458072 | 4.75 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_-_45956822 | 4.69 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr16_-_21436459 | 4.62 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr4_+_6784401 | 4.35 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr9_-_130712995 | 4.35 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr16_-_29517141 | 4.34 |
ENST00000550665.1
|
RP11-231C14.4
|
Uncharacterized protein |
| chr19_+_54641444 | 4.30 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr1_+_110754094 | 4.16 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr4_+_72052964 | 4.10 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr11_-_2182388 | 4.10 |
ENST00000421783.1
ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS
INS-IGF2
|
insulin INS-IGF2 readthrough |
| chr7_-_30029367 | 3.93 |
ENST00000242059.5
|
SCRN1
|
secernin 1 |
| chr19_+_12949251 | 3.88 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chrX_-_65858865 | 3.86 |
ENST00000374719.3
ENST00000450752.1 ENST00000451436.2 |
EDA2R
|
ectodysplasin A2 receptor |
| chr7_-_30029574 | 3.79 |
ENST00000426154.1
ENST00000421434.1 ENST00000434476.2 |
SCRN1
|
secernin 1 |
| chr4_+_72053017 | 3.71 |
ENST00000351898.6
|
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_-_54872059 | 3.62 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr5_+_176853702 | 3.40 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr2_-_30143525 | 3.37 |
ENST00000431873.1
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr16_-_29415350 | 3.22 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr2_+_26915584 | 2.99 |
ENST00000302909.3
|
KCNK3
|
potassium channel, subfamily K, member 3 |
| chrX_-_65859096 | 2.96 |
ENST00000456230.2
|
EDA2R
|
ectodysplasin A2 receptor |
| chr12_+_53817633 | 2.91 |
ENST00000257863.4
ENST00000550311.1 ENST00000379791.3 |
AMHR2
|
anti-Mullerian hormone receptor, type II |
| chr16_+_22524844 | 2.81 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr8_-_63998590 | 2.55 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chrX_-_106959631 | 2.46 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr11_-_62783303 | 2.44 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr11_-_62783276 | 2.21 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr1_+_200842083 | 2.20 |
ENST00000304244.2
|
GPR25
|
G protein-coupled receptor 25 |
| chr16_-_21868739 | 2.05 |
ENST00000415645.2
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr11_+_72929402 | 1.81 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr10_-_49701686 | 1.81 |
ENST00000417247.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr22_-_27014043 | 1.80 |
ENST00000215939.2
|
CRYBB1
|
crystallin, beta B1 |
| chr6_+_42896865 | 1.73 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr1_+_174769006 | 1.69 |
ENST00000489615.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr5_-_22853429 | 1.65 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr17_+_25936862 | 1.64 |
ENST00000582410.1
|
KSR1
|
kinase suppressor of ras 1 |
| chr11_-_66103932 | 1.60 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr5_-_39270725 | 1.58 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr9_+_131549610 | 1.50 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr11_+_118826999 | 1.49 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr6_+_7727030 | 1.43 |
ENST00000283147.6
|
BMP6
|
bone morphogenetic protein 6 |
| chr19_-_48547294 | 1.39 |
ENST00000293255.2
|
CABP5
|
calcium binding protein 5 |
| chr14_+_81421355 | 1.38 |
ENST00000541158.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chr11_+_72929319 | 1.37 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr7_-_22234381 | 1.30 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr19_+_36103631 | 1.28 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr1_+_202091980 | 1.22 |
ENST00000367282.5
|
GPR37L1
|
G protein-coupled receptor 37 like 1 |
| chr16_-_67514982 | 1.21 |
ENST00000565835.1
ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr10_-_29811456 | 1.08 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr10_-_70231639 | 1.03 |
ENST00000551118.2
ENST00000358410.3 ENST00000399180.2 ENST00000399179.2 |
DNA2
|
DNA replication helicase/nuclease 2 |
| chr7_-_100860851 | 0.89 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr10_-_81708854 | 0.72 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr12_-_22487618 | 0.72 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr2_+_85811525 | 0.71 |
ENST00000306384.4
|
VAMP5
|
vesicle-associated membrane protein 5 |
| chr10_-_79397479 | 0.64 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_+_54943134 | 0.59 |
ENST00000243052.3
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent |
| chr1_-_45956800 | 0.57 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr9_-_127905736 | 0.54 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr15_-_41047421 | 0.44 |
ENST00000560460.1
ENST00000338376.3 ENST00000560905.1 |
RMDN3
|
regulator of microtubule dynamics 3 |
| chr14_+_81421710 | 0.37 |
ENST00000342443.6
|
TSHR
|
thyroid stimulating hormone receptor |
| chr6_+_89791507 | 0.31 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_+_103236004 | 0.31 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr14_+_81421861 | 0.31 |
ENST00000298171.2
|
TSHR
|
thyroid stimulating hormone receptor |
| chrX_+_68725084 | 0.30 |
ENST00000252338.4
|
FAM155B
|
family with sequence similarity 155, member B |
| chr7_-_151433342 | 0.22 |
ENST00000433631.2
|
PRKAG2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr10_-_102289611 | 0.18 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr2_+_102624977 | 0.17 |
ENST00000441002.1
|
IL1R2
|
interleukin 1 receptor, type II |
| chrX_-_153141302 | 0.11 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr14_+_81421921 | 0.11 |
ENST00000554263.1
ENST00000554435.1 |
TSHR
|
thyroid stimulating hormone receptor |
| chr14_+_50779071 | 0.09 |
ENST00000426751.2
|
ATP5S
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr7_-_150754935 | 0.06 |
ENST00000297518.4
|
CDK5
|
cyclin-dependent kinase 5 |
| chr22_+_22599189 | 0.02 |
ENST00000302273.2
|
VPREB1
|
pre-B lymphocyte 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MAFA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.1 | 36.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 4.5 | 13.5 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 2.6 | 13.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.8 | 7.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 1.7 | 10.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 1.4 | 4.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 1.3 | 6.7 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.1 | 12.1 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.8 | 3.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.8 | 5.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.7 | 2.9 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.7 | 6.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.6 | 2.5 | GO:0090212 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.6 | 6.6 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.5 | 2.2 | GO:1905229 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.5 | 3.6 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.5 | 5.4 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 5.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.5 | 6.8 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.5 | 10.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.5 | 9.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.4 | 4.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 1.4 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.4 | 2.5 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.3 | 1.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 12.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 15.1 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.3 | 3.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.3 | 0.9 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.3 | 3.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 4.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 0.7 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 6.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 19.7 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.2 | 7.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.2 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 4.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 14.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 4.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 0.6 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.1 | 11.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 5.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 13.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 1.7 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 1.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 4.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 10.0 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 3.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 5.3 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.7 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 3.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 1.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.3 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.2 | GO:0046320 | regulation of fatty acid oxidation(GO:0046320) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.1 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) |
| 0.0 | 7.2 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 8.6 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 3.9 | GO:0007010 | cytoskeleton organization(GO:0007010) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 36.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.8 | 7.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.3 | 5.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 4.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 19.7 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 15.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 4.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 15.6 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 7.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 1.0 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 6.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 4.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 10.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 6.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 11.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 6.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.7 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 14.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 13.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 5.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 8.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 10.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 8.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 21.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 36.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 4.5 | 13.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 3.3 | 9.8 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 1.7 | 12.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.5 | 6.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 1.4 | 15.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 1.2 | 19.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.9 | 13.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.8 | 5.4 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.7 | 5.8 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.7 | 2.9 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.7 | 5.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.6 | 7.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.6 | 2.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.5 | 3.4 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.5 | 11.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 7.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 12.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 13.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.3 | 1.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 4.7 | GO:0015301 | inorganic anion exchanger activity(GO:0005452) anion:anion antiporter activity(GO:0015301) |
| 0.3 | 2.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.3 | 3.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 7.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 0.9 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 4.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 2.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.2 | 6.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 1.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 3.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 4.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 4.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 3.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 6.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 3.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 11.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 7.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 10.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 5.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.7 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 15.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.7 | 36.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 6.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 2.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 11.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 13.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 5.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 43.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.5 | 15.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.4 | 3.0 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.4 | 9.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.4 | 12.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 4.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 4.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 6.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 6.6 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 3.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 1.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 4.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 8.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 5.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 3.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |