Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for MECOM
Z-value: 1.00
Transcription factors associated with MECOM
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MECOM
|
ENSG00000085276.13 | MDS1 and EVI1 complex locus |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MECOM | hg19_v2_chr3_-_168865522_168865536, hg19_v2_chr3_-_169381183_169381258, hg19_v2_chr3_-_168864427_168864464 | -0.03 | 6.5e-01 | Click! |
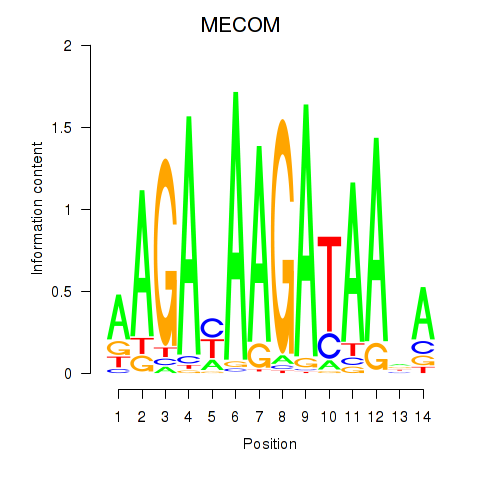
Activity profile of MECOM motif
Sorted Z-values of MECOM motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_88450244 | 33.21 |
ENST00000503414.1
|
SPARCL1
|
SPARC-like 1 (hevin) |
| chr12_-_16758059 | 19.10 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_16758304 | 18.74 |
ENST00000320122.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr6_-_31514516 | 16.95 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr17_-_66951474 | 16.25 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr8_-_27115931 | 15.13 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr6_-_52860171 | 13.61 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr12_-_15038779 | 13.15 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr6_-_52859968 | 12.97 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr10_+_106034884 | 12.43 |
ENST00000369707.2
ENST00000429569.2 |
GSTO2
|
glutathione S-transferase omega 2 |
| chr8_-_27115903 | 12.19 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr2_-_224467093 | 11.94 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr3_-_145968923 | 11.28 |
ENST00000493382.1
ENST00000354952.2 ENST00000383083.2 |
PLSCR4
|
phospholipid scramblase 4 |
| chr17_-_37308824 | 11.10 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr17_+_53344945 | 11.08 |
ENST00000575345.1
|
HLF
|
hepatic leukemia factor |
| chr3_-_145968857 | 10.61 |
ENST00000433593.2
ENST00000476202.1 ENST00000460885.1 |
PLSCR4
|
phospholipid scramblase 4 |
| chr5_+_140739537 | 10.32 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chrX_-_43741594 | 10.17 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr11_-_18656028 | 10.03 |
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr17_+_73750699 | 9.38 |
ENST00000584939.1
|
ITGB4
|
integrin, beta 4 |
| chr6_+_69942298 | 9.18 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr5_-_64920115 | 8.72 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr8_-_38008783 | 8.53 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr12_-_8218997 | 8.38 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr22_+_29138013 | 8.23 |
ENST00000216027.3
ENST00000398941.2 |
HSCB
|
HscB mitochondrial iron-sulfur cluster co-chaperone |
| chr18_-_21891460 | 8.17 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr7_+_97361388 | 8.17 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr3_-_45883558 | 8.03 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr2_+_189839046 | 7.92 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr2_+_55746746 | 7.01 |
ENST00000406691.3
ENST00000349456.4 ENST00000407816.3 ENST00000403007.3 |
CCDC104
|
coiled-coil domain containing 104 |
| chr7_-_100844193 | 6.95 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr4_-_76944621 | 6.91 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr3_+_147127142 | 6.87 |
ENST00000282928.4
|
ZIC1
|
Zic family member 1 |
| chr14_-_76447494 | 6.75 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr2_-_237416181 | 6.70 |
ENST00000409907.3
|
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr5_+_140165876 | 6.68 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr2_-_237416071 | 6.52 |
ENST00000309507.5
ENST00000431676.2 |
IQCA1
|
IQ motif containing with AAA domain 1 |
| chr14_-_24584138 | 6.20 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr5_+_64920543 | 6.20 |
ENST00000399438.3
ENST00000510585.2 |
TRAPPC13
CTC-534A2.2
|
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr9_+_82186682 | 6.15 |
ENST00000376552.2
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr2_-_56150910 | 5.96 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr17_-_43138357 | 5.95 |
ENST00000342350.5
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr9_+_134378289 | 5.92 |
ENST00000423007.1
ENST00000404875.2 ENST00000441334.1 ENST00000341012.7 ENST00000372228.3 ENST00000402686.3 ENST00000419118.2 ENST00000541219.1 ENST00000354713.4 ENST00000418774.1 ENST00000415075.1 ENST00000448212.1 ENST00000430619.1 |
POMT1
|
protein-O-mannosyltransferase 1 |
| chr12_+_57522258 | 5.90 |
ENST00000553277.1
ENST00000243077.3 |
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr8_-_39695719 | 5.87 |
ENST00000347580.4
ENST00000379853.2 ENST00000521880.1 |
ADAM2
|
ADAM metallopeptidase domain 2 |
| chr14_+_29236269 | 5.87 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr12_+_20848486 | 5.78 |
ENST00000545102.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr6_+_31515337 | 5.67 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr8_-_39695764 | 5.63 |
ENST00000265708.4
|
ADAM2
|
ADAM metallopeptidase domain 2 |
| chr5_+_74807886 | 5.60 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr5_-_115910091 | 5.58 |
ENST00000257414.8
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr6_-_87804815 | 5.56 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr5_+_102201722 | 5.52 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_145275228 | 5.38 |
ENST00000427902.1
ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr4_+_155484155 | 5.32 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_+_74275057 | 5.26 |
ENST00000511370.1
|
ALB
|
albumin |
| chr5_+_125695805 | 5.17 |
ENST00000513040.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr19_+_22235279 | 5.16 |
ENST00000594363.1
ENST00000597927.1 ENST00000594947.1 |
ZNF257
|
zinc finger protein 257 |
| chr6_+_29624898 | 5.09 |
ENST00000396704.3
ENST00000483013.1 ENST00000490427.1 ENST00000416766.2 ENST00000376891.4 ENST00000376898.3 ENST00000396701.2 ENST00000494692.1 ENST00000431798.2 |
MOG
|
myelin oligodendrocyte glycoprotein |
| chr17_-_37309480 | 5.07 |
ENST00000539608.1
|
PLXDC1
|
plexin domain containing 1 |
| chr6_-_22297730 | 4.96 |
ENST00000306482.1
|
PRL
|
prolactin |
| chrX_-_62974941 | 4.95 |
ENST00000374872.1
ENST00000253401.6 ENST00000374870.4 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chrX_-_85302531 | 4.95 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr5_+_64920826 | 4.86 |
ENST00000438419.2
ENST00000231526.4 ENST00000505553.1 ENST00000545191.1 |
TRAPPC13
|
trafficking protein particle complex 13 |
| chr3_+_148583043 | 4.86 |
ENST00000296046.3
|
CPA3
|
carboxypeptidase A3 (mast cell) |
| chr6_-_154831779 | 4.84 |
ENST00000607772.1
|
CNKSR3
|
CNKSR family member 3 |
| chr9_+_82186872 | 4.78 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr15_-_37392086 | 4.64 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr1_+_209859510 | 4.60 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr5_+_102201509 | 4.57 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr12_+_20848282 | 4.56 |
ENST00000545604.1
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr11_+_12115543 | 4.47 |
ENST00000537344.1
ENST00000532179.1 ENST00000526065.1 |
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr12_+_20848377 | 4.43 |
ENST00000540354.1
ENST00000266509.2 ENST00000381552.1 |
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_72079842 | 4.32 |
ENST00000266673.5
ENST00000550524.1 |
TMEM19
|
transmembrane protein 19 |
| chr11_+_61008514 | 4.24 |
ENST00000312403.5
|
PGA5
|
pepsinogen 5, group I (pepsinogen A) |
| chr1_+_206223941 | 4.18 |
ENST00000367126.4
|
AVPR1B
|
arginine vasopressin receptor 1B |
| chr15_-_79103757 | 4.12 |
ENST00000388820.4
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr7_-_8302164 | 3.99 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr8_+_42552533 | 3.89 |
ENST00000289957.2
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr13_+_49280951 | 3.87 |
ENST00000282018.3
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr15_+_71185148 | 3.87 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr14_-_24551195 | 3.81 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr12_+_72080253 | 3.73 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr14_-_24551137 | 3.69 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chrX_+_102883620 | 3.67 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr7_-_8302207 | 3.67 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr2_+_172543967 | 3.66 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr13_+_111855414 | 3.66 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chrX_-_101694853 | 3.63 |
ENST00000372749.1
|
NXF2B
|
nuclear RNA export factor 2B |
| chr8_+_134203303 | 3.57 |
ENST00000519433.1
ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr5_+_102201430 | 3.57 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_-_82089405 | 3.52 |
ENST00000554211.1
|
RP11-799P8.1
|
RP11-799P8.1 |
| chr2_-_157189180 | 3.52 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr13_-_46626847 | 3.48 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr7_-_15601595 | 3.35 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr3_+_152552685 | 3.24 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr19_-_46366392 | 3.16 |
ENST00000598059.1
ENST00000594293.1 ENST00000245934.7 |
SYMPK
|
symplekin |
| chr9_+_33240157 | 3.16 |
ENST00000379721.3
|
SPINK4
|
serine peptidase inhibitor, Kazal type 4 |
| chr3_-_184971853 | 3.15 |
ENST00000231887.3
|
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr1_+_50569575 | 3.14 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr11_-_5207612 | 3.13 |
ENST00000380367.1
|
OR52A1
|
olfactory receptor, family 52, subfamily A, member 1 |
| chr15_-_48470558 | 3.09 |
ENST00000324324.7
|
MYEF2
|
myelin expression factor 2 |
| chrX_+_15525426 | 3.06 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chrX_+_101502170 | 3.02 |
ENST00000372757.1
|
NXF2
|
nuclear RNA export factor 2 |
| chr2_+_172544011 | 2.99 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr16_+_33204156 | 2.90 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr3_+_4344988 | 2.80 |
ENST00000358065.4
|
SETMAR
|
SET domain and mariner transposase fusion gene |
| chr12_+_96588279 | 2.74 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr1_+_160160283 | 2.74 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr13_-_113242439 | 2.72 |
ENST00000375669.3
ENST00000261965.3 |
TUBGCP3
|
tubulin, gamma complex associated protein 3 |
| chr19_-_14316980 | 2.71 |
ENST00000361434.3
ENST00000340736.6 |
LPHN1
|
latrophilin 1 |
| chr14_+_22458631 | 2.70 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr12_-_371994 | 2.65 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr2_+_172543919 | 2.62 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_+_12202774 | 2.56 |
ENST00000589718.1
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr4_+_96012614 | 2.45 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_+_22857048 | 2.44 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr8_-_133637624 | 2.36 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr14_-_69261310 | 2.31 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr11_+_30252549 | 2.28 |
ENST00000254122.3
ENST00000417547.1 |
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr12_-_102872317 | 2.26 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr10_+_57358750 | 2.25 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr19_-_41388657 | 2.23 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr17_-_32484313 | 2.21 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr1_-_146696901 | 2.16 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chrX_+_66764375 | 2.12 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr12_+_12202785 | 2.11 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr4_+_164415785 | 2.09 |
ENST00000513272.1
ENST00000513134.1 |
TMA16
|
translation machinery associated 16 homolog (S. cerevisiae) |
| chr1_+_160160346 | 2.05 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr2_+_108145913 | 2.02 |
ENST00000443205.1
|
AC096669.3
|
AC096669.3 |
| chr5_-_160973649 | 2.00 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr3_-_119379427 | 1.98 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr6_+_153019069 | 1.96 |
ENST00000532295.1
|
MYCT1
|
myc target 1 |
| chr19_-_20844368 | 1.95 |
ENST00000595094.1
ENST00000601440.1 ENST00000291750.6 |
CTC-513N18.7
ZNF626
|
CTC-513N18.7 zinc finger protein 626 |
| chr22_+_36113919 | 1.95 |
ENST00000249044.2
|
APOL5
|
apolipoprotein L, 5 |
| chr8_+_39759794 | 1.95 |
ENST00000518804.1
ENST00000519154.1 ENST00000522495.1 ENST00000522840.1 |
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr4_+_155484103 | 1.95 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr16_-_90096309 | 1.88 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chrX_+_2959512 | 1.88 |
ENST00000381127.1
|
ARSF
|
arylsulfatase F |
| chr9_-_13175823 | 1.85 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr11_+_30253410 | 1.78 |
ENST00000533718.1
|
FSHB
|
follicle stimulating hormone, beta polypeptide |
| chr1_+_15802594 | 1.75 |
ENST00000375910.3
|
CELA2B
|
chymotrypsin-like elastase family, member 2B |
| chr19_+_47538560 | 1.69 |
ENST00000439365.2
ENST00000594670.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr16_-_69419871 | 1.66 |
ENST00000603068.1
ENST00000254942.3 ENST00000567296.2 |
TERF2
|
telomeric repeat binding factor 2 |
| chr1_+_171060018 | 1.66 |
ENST00000367755.4
ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3
|
flavin containing monooxygenase 3 |
| chr2_+_172544182 | 1.60 |
ENST00000409197.1
ENST00000456808.1 ENST00000409317.1 ENST00000409773.1 ENST00000411953.1 ENST00000409453.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr12_-_81763127 | 1.60 |
ENST00000541017.1
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr5_-_127674883 | 1.55 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr17_-_46691990 | 1.53 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr1_+_172422026 | 1.50 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr12_-_8815299 | 1.49 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr11_+_60989688 | 1.48 |
ENST00000378149.3
ENST00000422676.2 |
PGA4
|
pepsinogen 4, group I (pepsinogen A) |
| chr20_+_36012051 | 1.46 |
ENST00000373567.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr1_-_208084729 | 1.40 |
ENST00000310833.7
ENST00000356522.4 |
CD34
|
CD34 molecule |
| chr3_+_44916098 | 1.39 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr3_-_178984759 | 1.35 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr20_+_36932521 | 1.31 |
ENST00000262865.4
|
BPI
|
bactericidal/permeability-increasing protein |
| chr12_-_81763184 | 1.30 |
ENST00000548670.1
ENST00000541570.2 ENST00000553058.1 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr18_-_67624160 | 1.21 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr9_+_39355699 | 1.20 |
ENST00000377647.3
|
SPATA31A1
|
SPATA31 subfamily A, member 1 |
| chr10_-_29923893 | 1.15 |
ENST00000355867.4
|
SVIL
|
supervillin |
| chr5_+_54320078 | 1.12 |
ENST00000231009.2
|
GZMK
|
granzyme K (granzyme 3; tryptase II) |
| chr2_+_172544294 | 1.04 |
ENST00000358002.6
ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr4_-_40859132 | 1.01 |
ENST00000543538.1
ENST00000502841.1 ENST00000504305.1 ENST00000513516.1 ENST00000510670.1 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr6_-_10882174 | 1.01 |
ENST00000379491.4
|
GCM2
|
glial cells missing homolog 2 (Drosophila) |
| chr12_+_15699286 | 0.99 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr16_-_3930724 | 0.98 |
ENST00000262367.5
|
CREBBP
|
CREB binding protein |
| chr19_-_9785743 | 0.96 |
ENST00000537617.1
ENST00000589542.1 ENST00000590155.1 ENST00000541032.1 ENST00000588653.1 ENST00000448622.1 ENST00000453792.2 |
ZNF562
|
zinc finger protein 562 |
| chr3_-_38992052 | 0.87 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr10_-_48806939 | 0.87 |
ENST00000374233.3
ENST00000507417.1 ENST00000512321.1 ENST00000395660.2 ENST00000374235.2 ENST00000395661.3 |
PTPN20B
|
protein tyrosine phosphatase, non-receptor type 20B |
| chr11_+_22696314 | 0.85 |
ENST00000532398.1
ENST00000433790.1 |
GAS2
|
growth arrest-specific 2 |
| chr21_-_28338732 | 0.80 |
ENST00000284987.5
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chrX_-_49965663 | 0.79 |
ENST00000376056.2
ENST00000376058.2 ENST00000358526.2 |
AKAP4
|
A kinase (PRKA) anchor protein 4 |
| chrX_+_30261847 | 0.79 |
ENST00000378981.3
ENST00000397550.1 |
MAGEB1
|
melanoma antigen family B, 1 |
| chr8_-_95274536 | 0.78 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr16_+_31225337 | 0.75 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr9_-_95244781 | 0.73 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chrX_+_155227246 | 0.69 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chr13_+_60971080 | 0.67 |
ENST00000377894.2
|
TDRD3
|
tudor domain containing 3 |
| chr11_-_8964580 | 0.67 |
ENST00000325884.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chrX_+_129473916 | 0.66 |
ENST00000545805.1
ENST00000543953.1 ENST00000218197.5 |
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_+_86934526 | 0.60 |
ENST00000394711.1
|
CLCA1
|
chloride channel accessory 1 |
| chr12_-_47219733 | 0.58 |
ENST00000547477.1
ENST00000447411.1 ENST00000266579.4 |
SLC38A4
|
solute carrier family 38, member 4 |
| chr8_-_93029865 | 0.57 |
ENST00000422361.2
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr19_-_47735918 | 0.57 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr1_+_47901689 | 0.53 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr5_+_92919043 | 0.53 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr12_+_41831485 | 0.47 |
ENST00000539469.2
ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr4_+_74301880 | 0.45 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr3_+_121311966 | 0.44 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr6_+_29141311 | 0.44 |
ENST00000377167.2
|
OR2J2
|
olfactory receptor, family 2, subfamily J, member 2 |
| chr10_-_69455873 | 0.34 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr5_+_140787600 | 0.32 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr1_+_82266053 | 0.25 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr9_-_107690420 | 0.22 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chrX_+_135730373 | 0.19 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr12_+_21168630 | 0.16 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr16_+_2867228 | 0.12 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr13_-_103053946 | 0.04 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chrX_-_10588595 | 0.04 |
ENST00000423614.1
ENST00000317552.4 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
Network of associatons between targets according to the STRING database.
First level regulatory network of MECOM
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 37.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 3.4 | 13.7 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 3.4 | 13.7 | GO:0031179 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 2.8 | 8.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 2.7 | 8.2 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 2.0 | 5.9 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 1.8 | 12.4 | GO:0090487 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 1.8 | 5.3 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 1.7 | 6.7 | GO:1905073 | ossification involved in bone remodeling(GO:0043932) frontal suture morphogenesis(GO:0060364) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.7 | 21.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 1.6 | 8.0 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 1.5 | 4.5 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 1.5 | 16.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.5 | 10.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 1.4 | 6.9 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 1.4 | 8.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.3 | 5.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 1.2 | 5.9 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 1.2 | 3.5 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 1.2 | 2.3 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 1.1 | 14.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.1 | 10.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 1.1 | 11.9 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 1.0 | 7.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.9 | 2.8 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.9 | 26.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.8 | 3.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.8 | 11.9 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.7 | 7.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.7 | 9.4 | GO:0035878 | nail development(GO:0035878) |
| 0.7 | 2.1 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.7 | 4.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.7 | 4.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.7 | 6.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.6 | 3.9 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.6 | 11.5 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.6 | 15.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.6 | 4.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.6 | 8.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.6 | 1.7 | GO:0031627 | telomeric loop formation(GO:0031627) negative regulation of t-circle formation(GO:1904430) |
| 0.5 | 4.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.5 | 26.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.5 | 17.0 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.5 | 4.9 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.5 | 1.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.5 | 3.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 5.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.5 | 2.3 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.4 | 6.9 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.4 | 1.2 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.4 | 2.7 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.4 | 2.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.4 | 3.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.4 | 1.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.3 | 1.4 | GO:1900168 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.3 | 5.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 | 1.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 2.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 3.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.3 | 2.4 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.3 | 4.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.3 | 5.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 0.8 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 | 0.7 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.2 | 24.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.2 | 2.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 1.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 4.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 3.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 4.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.2 | 5.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 8.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.2 | 5.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 5.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.2 | 2.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 1.0 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 11.1 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 3.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.3 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.1 | 0.7 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.1 | 2.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 17.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 5.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 2.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 3.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.7 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.1 | 0.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 3.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 3.3 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 3.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 28.7 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.1 | 0.3 | GO:0086073 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.2 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 2.9 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.9 | GO:0044764 | viral process(GO:0016032) multi-organism cellular process(GO:0044764) |
| 0.0 | 1.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 5.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 3.1 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 5.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 7.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.9 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 1.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.6 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 2.0 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.0 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle recycling(GO:1903421) |
| 0.0 | 0.6 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 2.4 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 1.2 | 8.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.7 | 17.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.7 | 7.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 11.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 3.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.6 | 4.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.6 | 5.7 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.6 | 7.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.5 | 2.7 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 1.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.5 | 3.7 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.5 | 3.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.4 | 9.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 11.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.3 | 2.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 1.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 2.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 3.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 62.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 29.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 5.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 18.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 11.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 8.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 2.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 13.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 5.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 5.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 2.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 4.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 13.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 36.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 3.4 | 13.7 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 2.3 | 6.9 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 2.2 | 6.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 2.0 | 16.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 2.0 | 21.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.8 | 14.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 1.7 | 5.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 1.5 | 4.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.5 | 6.1 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) |
| 1.5 | 5.9 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 1.3 | 6.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.3 | 10.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.2 | 6.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.0 | 4.9 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 1.0 | 5.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.9 | 2.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.9 | 10.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.8 | 3.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.8 | 3.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.8 | 3.9 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.8 | 13.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.7 | 9.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.6 | 1.9 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.6 | 8.4 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.6 | 11.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.6 | 7.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 16.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.5 | 34.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.5 | 3.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.5 | 2.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.5 | 4.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.4 | 2.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.4 | 2.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.4 | 2.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.4 | 3.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 11.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 5.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 14.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.3 | 2.0 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.3 | 2.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 5.7 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 8.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 1.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 8.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 2.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 11.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 5.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 3.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.2 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 14.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 1.7 | GO:0034061 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) DNA polymerase activity(GO:0034061) |
| 0.2 | 2.0 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.2 | 4.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 4.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.2 | 5.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 16.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.4 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 1.9 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 2.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 17.0 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 0.1 | 5.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 11.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 2.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 3.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 8.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 7.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 27.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 3.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 15.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 13.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 60.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 5.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 6.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 7.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 8.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 21.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 9.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 5.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 12.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.9 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 3.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 5.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 8.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 39.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.9 | 20.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.7 | 9.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.6 | 17.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.4 | 16.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 6.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 13.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 5.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 3.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 7.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.9 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 5.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 3.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 10.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 2.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 11.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 1.9 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 27.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 18.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 7.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 10.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 6.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 2.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 6.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |