Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
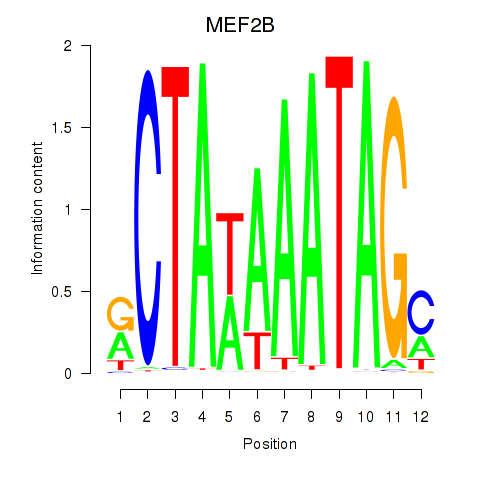
Results for MEF2B
Z-value: 0.64
Transcription factors associated with MEF2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2B
|
ENSG00000213999.11 | myocyte enhancer factor 2B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2B | hg19_v2_chr19_-_19302931_19302974 | 0.28 | 3.1e-05 | Click! |
Activity profile of MEF2B motif
Sorted Z-values of MEF2B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_21493884 | 22.18 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr14_-_21493649 | 21.23 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr20_+_10199468 | 20.99 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr14_-_21493123 | 20.46 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr8_-_27115931 | 19.21 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chr2_+_233497931 | 19.09 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr8_-_27115903 | 17.15 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr12_-_45269430 | 15.79 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr12_-_49393092 | 13.57 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr20_+_44035847 | 13.53 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr3_+_127634069 | 10.40 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr3_+_127634312 | 10.31 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr5_-_42811986 | 10.15 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr20_+_44657845 | 9.79 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr5_-_42812143 | 9.42 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr7_-_37488834 | 9.31 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_-_86564776 | 9.30 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr12_+_52445191 | 8.84 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_+_84328252 | 8.66 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr16_+_84328429 | 8.42 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr1_+_92632542 | 7.80 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr12_+_119616447 | 7.71 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr15_+_42651691 | 7.35 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr14_-_22005343 | 7.20 |
ENST00000327430.3
|
SALL2
|
spalt-like transcription factor 2 |
| chr11_-_18034701 | 6.46 |
ENST00000265965.5
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr3_+_138067521 | 6.35 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr2_-_157198860 | 6.26 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr3_+_138067314 | 6.19 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr8_+_95565947 | 5.83 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chr22_+_32340447 | 5.71 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr22_+_32340481 | 5.63 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr1_+_84630645 | 5.52 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_+_86781847 | 5.46 |
ENST00000432366.2
ENST00000423590.2 ENST00000394703.5 |
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr17_+_20059302 | 5.45 |
ENST00000395530.2
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_-_19229014 | 5.20 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr5_-_16509101 | 5.14 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr17_+_7184986 | 4.75 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr3_+_138067666 | 4.56 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr2_+_33661382 | 4.53 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr8_+_107282389 | 4.29 |
ENST00000577661.1
ENST00000445937.1 |
RP11-395G23.3
OXR1
|
RP11-395G23.3 oxidation resistance 1 |
| chr1_+_77333117 | 4.07 |
ENST00000477717.1
|
ST6GALNAC5
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr4_+_41362796 | 3.93 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr9_+_100263912 | 3.74 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr5_-_131347501 | 3.50 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr7_+_30951461 | 3.20 |
ENST00000311813.4
|
AQP1
|
aquaporin 1 (Colton blood group) |
| chr3_+_159570722 | 3.07 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_-_371994 | 3.04 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr3_+_186739636 | 2.99 |
ENST00000440338.1
ENST00000448044.1 |
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_-_188665428 | 2.90 |
ENST00000444488.1
|
TPRG1-AS1
|
TPRG1 antisense RNA 1 |
| chr1_-_156460391 | 2.83 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr12_+_25205446 | 2.79 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr5_-_131347583 | 2.64 |
ENST00000379255.1
ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr15_+_40531243 | 2.63 |
ENST00000558055.1
ENST00000455577.2 |
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr21_-_42219065 | 2.61 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr3_-_192445289 | 2.47 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr12_+_25205666 | 2.38 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr12_+_15699286 | 2.37 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr15_+_100106126 | 2.26 |
ENST00000558812.1
ENST00000338042.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr12_+_25205568 | 2.25 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr17_-_42200996 | 2.25 |
ENST00000587135.1
ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5
|
histone deacetylase 5 |
| chr15_+_100106155 | 2.06 |
ENST00000557785.1
ENST00000558049.1 ENST00000449277.2 |
MEF2A
|
myocyte enhancer factor 2A |
| chr6_+_139094657 | 2.03 |
ENST00000332797.6
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr8_+_1993152 | 2.00 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr17_-_42200958 | 1.95 |
ENST00000336057.5
|
HDAC5
|
histone deacetylase 5 |
| chr20_+_11898507 | 1.94 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr3_+_54157480 | 1.85 |
ENST00000490478.1
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr19_-_40324255 | 1.76 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr1_+_26348259 | 1.55 |
ENST00000374280.3
|
EXTL1
|
exostosin-like glycosyltransferase 1 |
| chr5_+_53751445 | 1.55 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr1_+_160051319 | 1.46 |
ENST00000368088.3
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr11_-_47400078 | 1.45 |
ENST00000378538.3
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr22_-_51017084 | 1.44 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr4_+_120056939 | 1.43 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr11_+_1860682 | 1.42 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr7_+_143013198 | 1.40 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr15_+_40531621 | 1.38 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr1_-_116311402 | 1.36 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr18_+_12407895 | 1.26 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr19_-_40324767 | 1.21 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr1_+_22778337 | 1.20 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr1_-_27240455 | 1.19 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr5_-_131347306 | 1.15 |
ENST00000296869.4
ENST00000379249.3 ENST00000379272.2 ENST00000379264.2 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr6_-_169654139 | 1.15 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr10_-_75415825 | 1.15 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr8_-_70745575 | 1.13 |
ENST00000524945.1
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1 |
| chr11_+_1860832 | 1.07 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr11_-_47374246 | 1.02 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr16_+_12058961 | 0.93 |
ENST00000053243.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr22_-_51016846 | 0.92 |
ENST00000312108.7
ENST00000395650.2 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr8_+_1993173 | 0.90 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr8_+_11351876 | 0.88 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr6_-_33160231 | 0.85 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr11_-_84634447 | 0.84 |
ENST00000532653.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr15_+_100106244 | 0.83 |
ENST00000557942.1
|
MEF2A
|
myocyte enhancer factor 2A |
| chr3_+_8543533 | 0.80 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr16_-_31439735 | 0.79 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr17_-_34417479 | 0.78 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr10_-_97200772 | 0.73 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr11_-_84634217 | 0.72 |
ENST00000524982.1
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr16_+_12059050 | 0.71 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chrX_-_11284095 | 0.62 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr17_-_1389228 | 0.58 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr10_+_135340859 | 0.55 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr5_-_66492562 | 0.49 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr16_+_12059091 | 0.48 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr11_-_47399942 | 0.46 |
ENST00000227163.4
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_+_11351494 | 0.46 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr2_+_170366203 | 0.45 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr15_+_68346501 | 0.45 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr16_+_31225337 | 0.38 |
ENST00000322122.3
|
TRIM72
|
tripartite motif containing 72 |
| chr11_+_7597639 | 0.33 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_-_113559104 | 0.19 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr6_+_152128371 | 0.12 |
ENST00000443427.1
|
ESR1
|
estrogen receptor 1 |
| chr5_+_138609782 | 0.08 |
ENST00000361059.2
ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3
|
matrin 3 |
| chr5_+_132009675 | 0.02 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 21.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 4.0 | 63.9 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.4 | 9.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 2.1 | 6.3 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 1.3 | 5.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 1.3 | 8.8 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.2 | 7.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 1.1 | 15.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.1 | 5.5 | GO:0097338 | response to clozapine(GO:0097338) |
| 1.1 | 3.2 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.9 | 5.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.8 | 2.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.7 | 36.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.6 | 3.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.6 | 9.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.6 | 11.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 1.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 7.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.3 | 7.1 | GO:0097503 | sialylation(GO:0097503) |
| 0.3 | 4.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 2.5 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 9.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 0.8 | GO:0060023 | soft palate development(GO:0060023) |
| 0.2 | 2.8 | GO:0007512 | adult heart development(GO:0007512) |
| 0.2 | 2.6 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 3.9 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.2 | 3.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 | 17.1 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.2 | 4.8 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.1 | 1.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 2.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 17.1 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.6 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.5 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 7.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 1.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 3.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.4 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 1.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.5 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 7.2 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 1.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 2.1 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.7 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 4.5 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 3.3 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 2.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 22.7 | GO:0031175 | neuron projection development(GO:0031175) |
| 0.0 | 4.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 1.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 12.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 1.0 | GO:0030282 | bone mineralization(GO:0030282) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 21.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.2 | 13.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.1 | 3.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.6 | 13.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 4.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.5 | 105.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.3 | 1.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 21.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 5.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 9.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 7.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 13.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 2.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 7.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 10.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 6.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 7.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 7.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 3.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 7.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 5.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 2.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.1 | 3.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 1.0 | 4.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 1.0 | 3.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.9 | 6.5 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.9 | 9.8 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.7 | 17.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.7 | 21.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.6 | 8.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.5 | 3.0 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.5 | 17.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.5 | 1.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 2.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.4 | 7.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.3 | 5.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 4.8 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.3 | 2.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 1.5 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 4.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 5.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 17.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.8 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 1.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 3.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 5.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.8 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 37.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 8.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 4.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 21.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 9.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.8 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 65.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.4 | 16.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 8.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 9.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.2 | 5.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 19.7 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 5.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 4.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 4.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 16.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.2 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 21.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 7.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 3.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 5.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 9.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.2 | 8.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 2.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 4.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 8.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 3.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 9.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 7.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.1 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 4.5 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.3 | REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | Genes involved in Signaling by the B Cell Receptor (BCR) |
| 0.0 | 5.2 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |