Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
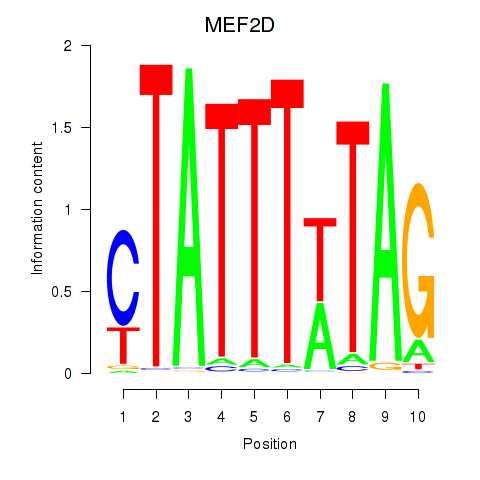
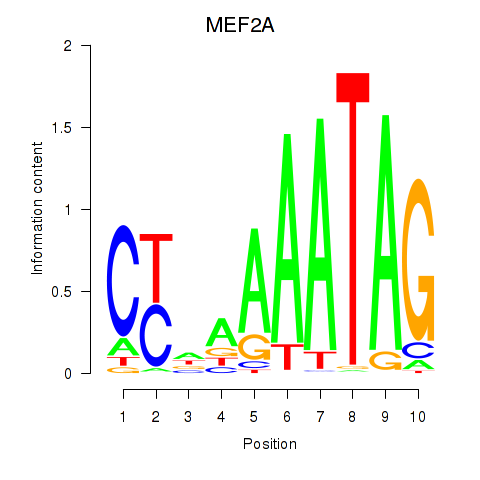
Results for MEF2D_MEF2A
Z-value: 1.27
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2D | hg19_v2_chr1_-_156470515_156470542 | 0.53 | 4.7e-17 | Click! |
| MEF2A | hg19_v2_chr15_+_100106155_100106243 | 0.21 | 1.4e-03 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_21493884 | 40.13 |
ENST00000556974.1
ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2
|
NDRG family member 2 |
| chr12_-_45269430 | 39.39 |
ENST00000395487.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr14_-_21493649 | 36.59 |
ENST00000553442.1
ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2
|
NDRG family member 2 |
| chr14_-_21493123 | 36.03 |
ENST00000556147.1
ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2
|
NDRG family member 2 |
| chr2_+_233497931 | 32.11 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr3_+_127634312 | 29.46 |
ENST00000407609.3
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr3_+_127634069 | 26.95 |
ENST00000405109.1
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr12_-_111358372 | 26.92 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr11_-_19223523 | 25.43 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr11_+_1860200 | 25.15 |
ENST00000381911.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr2_-_211168332 | 24.47 |
ENST00000341685.4
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr6_-_6007200 | 24.29 |
ENST00000244766.2
|
NRN1
|
neuritin 1 |
| chr1_+_160085501 | 22.75 |
ENST00000361216.3
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr3_+_138067521 | 20.65 |
ENST00000494949.1
|
MRAS
|
muscle RAS oncogene homolog |
| chr10_+_95517660 | 20.61 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr16_-_31439735 | 19.91 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr19_+_35629702 | 19.75 |
ENST00000351325.4
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr20_+_44657845 | 19.50 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr9_+_139871948 | 19.42 |
ENST00000224167.2
ENST00000457950.1 ENST00000371625.3 ENST00000371623.3 |
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr13_-_67802549 | 19.15 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr19_+_35630022 | 18.74 |
ENST00000589209.1
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr3_+_138067314 | 17.70 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr12_-_91539918 | 17.68 |
ENST00000548218.1
|
DCN
|
decorin |
| chr22_-_36013368 | 17.67 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr8_-_27115931 | 17.44 |
ENST00000523048.1
|
STMN4
|
stathmin-like 4 |
| chrX_-_21776281 | 16.35 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr15_+_42651691 | 16.33 |
ENST00000357568.3
ENST00000349748.3 ENST00000318023.7 ENST00000397163.3 |
CAPN3
|
calpain 3, (p94) |
| chr10_+_88428370 | 16.19 |
ENST00000372066.3
ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3
|
LIM domain binding 3 |
| chr10_+_88428206 | 16.10 |
ENST00000429277.2
ENST00000458213.2 ENST00000352360.5 |
LDB3
|
LIM domain binding 3 |
| chr2_-_211179883 | 15.99 |
ENST00000352451.3
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr12_+_119616447 | 15.56 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr8_+_1993152 | 15.34 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr17_+_37821593 | 15.28 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr7_+_30951461 | 15.27 |
ENST00000311813.4
|
AQP1
|
aquaporin 1 (Colton blood group) |
| chr10_-_75415825 | 15.10 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like |
| chr10_+_95517616 | 15.09 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr19_-_47164386 | 15.00 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr8_-_27115903 | 14.96 |
ENST00000350889.4
ENST00000519997.1 ENST00000519614.1 ENST00000522908.1 ENST00000265770.7 |
STMN4
|
stathmin-like 4 |
| chr12_-_16759711 | 14.86 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_138067666 | 14.85 |
ENST00000475711.1
ENST00000464896.1 |
MRAS
|
muscle RAS oncogene homolog |
| chr5_-_42812143 | 14.60 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_+_1860832 | 14.47 |
ENST00000252898.7
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr5_-_42811986 | 14.47 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr20_+_44035847 | 14.28 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr14_-_21491477 | 14.13 |
ENST00000298684.5
ENST00000557169.1 ENST00000553563.1 |
NDRG2
|
NDRG family member 2 |
| chr7_-_37488834 | 13.98 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_+_1860682 | 13.96 |
ENST00000381906.1
|
TNNI2
|
troponin I type 2 (skeletal, fast) |
| chr9_+_100263912 | 13.94 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr4_+_41362796 | 13.67 |
ENST00000508501.1
ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr8_+_27631903 | 13.63 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_-_49393092 | 13.50 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr12_-_9268707 | 13.31 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr1_-_201391149 | 13.26 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr22_-_36018569 | 13.18 |
ENST00000419229.1
ENST00000406324.1 |
MB
|
myoglobin |
| chr16_+_7382745 | 13.08 |
ENST00000436368.2
ENST00000311745.5 ENST00000355637.4 ENST00000340209.4 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr7_-_44105158 | 12.52 |
ENST00000297283.3
|
PGAM2
|
phosphoglycerate mutase 2 (muscle) |
| chr17_+_7184986 | 12.42 |
ENST00000317370.8
ENST00000571308.1 |
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr10_+_95517566 | 12.40 |
ENST00000542308.1
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr17_+_4855053 | 12.17 |
ENST00000518175.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr8_+_1993173 | 12.12 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr16_+_8768422 | 12.03 |
ENST00000268251.8
ENST00000564714.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr5_+_53751445 | 11.51 |
ENST00000302005.1
|
HSPB3
|
heat shock 27kDa protein 3 |
| chr11_+_1940925 | 11.43 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr4_+_186064395 | 11.39 |
ENST00000281456.6
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr9_+_129677039 | 11.21 |
ENST00000259351.5
ENST00000424082.2 ENST00000394022.3 ENST00000394011.3 ENST00000319107.4 |
RALGPS1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr12_+_52445191 | 10.86 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr12_+_101988627 | 10.64 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr4_+_113970772 | 10.57 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr1_-_154164534 | 10.40 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chr10_-_115423792 | 10.37 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr22_+_32340481 | 10.36 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr14_-_23877474 | 10.28 |
ENST00000405093.3
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr3_-_52486841 | 10.11 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr11_+_1942580 | 10.11 |
ENST00000381558.1
|
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr11_-_47374246 | 9.93 |
ENST00000545968.1
ENST00000399249.2 ENST00000256993.4 |
MYBPC3
|
myosin binding protein C, cardiac |
| chr11_+_112832202 | 9.83 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr2_+_220283091 | 9.63 |
ENST00000373960.3
|
DES
|
desmin |
| chr1_-_31845914 | 9.52 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_+_120056939 | 9.34 |
ENST00000307128.5
|
MYOZ2
|
myozenin 2 |
| chr14_+_100150622 | 9.26 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr11_+_112832090 | 9.20 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr5_+_67584174 | 9.18 |
ENST00000320694.8
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_+_66300446 | 9.14 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_116311402 | 9.02 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr1_-_115238207 | 8.87 |
ENST00000520113.2
ENST00000369538.3 ENST00000353928.6 |
AMPD1
|
adenosine monophosphate deaminase 1 |
| chr1_+_92632542 | 8.79 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr11_-_18034701 | 8.64 |
ENST00000265965.5
|
SERGEF
|
secretion regulating guanine nucleotide exchange factor |
| chr9_-_97356075 | 8.61 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr6_+_7108210 | 8.58 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr5_+_161274940 | 8.55 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr12_-_45270151 | 8.52 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr16_+_6069072 | 8.49 |
ENST00000547605.1
ENST00000550418.1 ENST00000553186.1 |
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr22_+_32340447 | 8.36 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr11_-_62477041 | 8.33 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr12_-_45270077 | 8.30 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr5_-_88179302 | 8.26 |
ENST00000504921.2
|
MEF2C
|
myocyte enhancer factor 2C |
| chr7_-_150946015 | 8.22 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr1_+_160160283 | 8.22 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr19_+_37998031 | 8.22 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr4_-_57547870 | 8.21 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr19_+_3933579 | 8.18 |
ENST00000593949.1
|
NMRK2
|
nicotinamide riboside kinase 2 |
| chr5_+_161274685 | 8.16 |
ENST00000428797.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr16_+_84328252 | 8.13 |
ENST00000219454.5
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr17_+_42264322 | 8.07 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr19_+_35630344 | 8.05 |
ENST00000455515.2
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr4_-_57547454 | 7.79 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr16_+_84328429 | 7.73 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr19_-_47975417 | 7.60 |
ENST00000236877.6
|
SLC8A2
|
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr2_+_33661382 | 7.59 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr2_+_220143989 | 7.52 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr8_-_17533838 | 7.52 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr4_-_186697044 | 7.40 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr19_+_3933085 | 7.40 |
ENST00000168977.2
ENST00000599576.1 |
NMRK2
|
nicotinamide riboside kinase 2 |
| chr7_+_47694842 | 7.32 |
ENST00000408988.2
|
C7orf65
|
chromosome 7 open reading frame 65 |
| chr1_+_160160346 | 7.30 |
ENST00000368078.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr6_-_169654139 | 7.22 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr7_+_99775520 | 7.15 |
ENST00000317296.5
ENST00000422690.1 ENST00000439782.1 |
STAG3
|
stromal antigen 3 |
| chr18_-_53070913 | 6.97 |
ENST00000568186.1
ENST00000564228.1 |
TCF4
|
transcription factor 4 |
| chr7_+_99775366 | 6.70 |
ENST00000394018.2
ENST00000416412.1 |
STAG3
|
stromal antigen 3 |
| chr1_-_201390846 | 6.65 |
ENST00000367312.1
ENST00000555340.2 ENST00000361379.4 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr2_+_170366203 | 6.56 |
ENST00000284669.1
|
KLHL41
|
kelch-like family member 41 |
| chr7_+_154002189 | 6.49 |
ENST00000332007.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr2_+_173686303 | 6.37 |
ENST00000397087.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chrX_+_153046456 | 6.36 |
ENST00000393786.3
ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3
|
SRSF protein kinase 3 |
| chr17_-_1389228 | 6.24 |
ENST00000438665.2
|
MYO1C
|
myosin IC |
| chr7_-_134143841 | 6.15 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr12_+_12938541 | 6.00 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr22_+_31742875 | 5.96 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr6_-_27806117 | 5.95 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr5_-_115910630 | 5.94 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_-_117753540 | 5.92 |
ENST00000328189.3
ENST00000369458.3 |
VTCN1
|
V-set domain containing T cell activation inhibitor 1 |
| chr18_-_3220106 | 5.91 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr22_-_37215523 | 5.78 |
ENST00000216200.5
|
PVALB
|
parvalbumin |
| chr11_+_1940786 | 5.77 |
ENST00000278317.6
ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr17_-_9929581 | 5.75 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr5_-_137475071 | 5.74 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr15_+_73976715 | 5.73 |
ENST00000558689.1
ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276
|
CD276 molecule |
| chr3_-_38071122 | 5.63 |
ENST00000334661.4
|
PLCD1
|
phospholipase C, delta 1 |
| chr7_-_113559104 | 5.63 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr12_+_69201923 | 5.61 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr9_+_33290491 | 5.60 |
ENST00000379540.3
ENST00000379521.4 ENST00000318524.6 |
NFX1
|
nuclear transcription factor, X-box binding 1 |
| chr5_-_16509101 | 5.47 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr1_+_95583479 | 5.44 |
ENST00000455656.1
ENST00000604534.1 |
TMEM56
RP11-57H12.6
|
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr1_+_150229554 | 5.34 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr17_-_10421853 | 5.27 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chr3_-_46904918 | 5.19 |
ENST00000395869.1
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr8_+_104831472 | 5.16 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr3_-_192445289 | 5.15 |
ENST00000430714.1
ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12
|
fibroblast growth factor 12 |
| chr2_+_30569506 | 5.14 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr3_-_46904946 | 4.98 |
ENST00000292327.4
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow |
| chr12_-_371994 | 4.97 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr9_-_130639997 | 4.94 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr1_-_201346761 | 4.93 |
ENST00000455702.1
ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2
|
troponin T type 2 (cardiac) |
| chr7_+_143013198 | 4.74 |
ENST00000343257.2
|
CLCN1
|
chloride channel, voltage-sensitive 1 |
| chr4_-_186696425 | 4.69 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_156460391 | 4.68 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr5_+_140552218 | 4.67 |
ENST00000231137.3
|
PCDHB7
|
protocadherin beta 7 |
| chr15_+_91446157 | 4.63 |
ENST00000559717.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr10_-_45474237 | 4.61 |
ENST00000448778.1
ENST00000298295.3 |
C10orf10
|
chromosome 10 open reading frame 10 |
| chr20_+_1875110 | 4.59 |
ENST00000400068.3
|
SIRPA
|
signal-regulatory protein alpha |
| chr1_-_25756638 | 4.55 |
ENST00000349320.3
|
RHCE
|
Rh blood group, CcEe antigens |
| chr18_+_13611431 | 4.54 |
ENST00000587757.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr11_+_45918092 | 4.53 |
ENST00000395629.2
|
MAPK8IP1
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr10_-_6104253 | 4.50 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr10_-_74114714 | 4.36 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr2_+_220144052 | 4.29 |
ENST00000425450.1
ENST00000392086.4 ENST00000421532.1 |
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr8_+_95565947 | 4.22 |
ENST00000523011.1
|
RP11-267M23.4
|
RP11-267M23.4 |
| chrX_-_11284095 | 4.19 |
ENST00000303025.6
ENST00000534860.1 |
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr5_-_131347501 | 4.16 |
ENST00000543479.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr17_+_68165657 | 4.16 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr7_-_151217001 | 4.13 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr6_+_41021027 | 4.13 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr3_+_35721106 | 4.09 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_+_26020672 | 3.93 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr10_-_72141330 | 3.91 |
ENST00000395011.1
ENST00000395010.1 |
LRRC20
|
leucine rich repeat containing 20 |
| chr11_-_35440796 | 3.87 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_-_104179682 | 3.86 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr17_+_42264556 | 3.84 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chrX_+_9217932 | 3.81 |
ENST00000432442.1
|
GS1-519E5.1
|
GS1-519E5.1 |
| chr11_-_35440579 | 3.76 |
ENST00000606205.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr9_-_215744 | 3.73 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr2_-_166060552 | 3.71 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chr2_+_113816685 | 3.70 |
ENST00000393200.2
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr22_-_31742218 | 3.68 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr9_-_136344197 | 3.62 |
ENST00000414172.1
ENST00000371897.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr3_-_114343039 | 3.55 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr10_-_75410771 | 3.49 |
ENST00000372873.4
|
SYNPO2L
|
synaptopodin 2-like |
| chr3_+_8543561 | 3.46 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr21_-_42219065 | 3.46 |
ENST00000400454.1
|
DSCAM
|
Down syndrome cell adhesion molecule |
| chr1_+_25757376 | 3.41 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr9_-_15472730 | 3.33 |
ENST00000481862.1
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr9_-_136344237 | 3.31 |
ENST00000432868.1
ENST00000371899.4 |
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr2_-_157198860 | 3.30 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr12_+_81101277 | 3.28 |
ENST00000228641.3
|
MYF6
|
myogenic factor 6 (herculin) |
| chr8_+_27183033 | 3.26 |
ENST00000420218.2
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr3_+_159570722 | 3.25 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr16_-_71610985 | 3.22 |
ENST00000355962.4
|
TAT
|
tyrosine aminotransferase |
| chr10_-_104178857 | 3.18 |
ENST00000020673.5
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr16_+_30075783 | 3.16 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chrX_+_119384607 | 3.11 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.2 | 40.7 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 7.9 | 126.9 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 7.8 | 46.5 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 5.6 | 5.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 5.1 | 15.3 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 5.0 | 25.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 4.9 | 19.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 4.5 | 22.7 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 4.0 | 56.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 3.9 | 30.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 3.8 | 30.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 3.5 | 10.6 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 3.4 | 26.9 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 3.2 | 9.5 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 3.1 | 12.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 3.1 | 15.5 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 3.0 | 12.0 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 2.7 | 208.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 2.7 | 16.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 2.5 | 14.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 2.2 | 8.6 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 2.1 | 8.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 2.1 | 8.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 2.1 | 6.2 | GO:0006059 | hexitol metabolic process(GO:0006059) response to methylglyoxal(GO:0051595) |
| 1.9 | 11.4 | GO:0015853 | adenine transport(GO:0015853) |
| 1.9 | 5.6 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 1.7 | 19.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.6 | 8.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.6 | 14.7 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 1.6 | 6.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 1.5 | 6.1 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 1.5 | 5.9 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.5 | 8.8 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 1.3 | 46.4 | GO:0045214 | sarcomere organization(GO:0045214) |
| 1.3 | 10.2 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 1.1 | 19.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.1 | 4.5 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.0 | 4.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 1.0 | 13.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.9 | 19.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.9 | 2.8 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.9 | 28.0 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.9 | 8.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.8 | 0.8 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.8 | 16.0 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.8 | 16.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 13.9 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.8 | 2.4 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.7 | 5.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.7 | 2.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.7 | 19.4 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.7 | 4.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.7 | 15.6 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.7 | 32.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.6 | 7.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.6 | 3.7 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.6 | 2.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.6 | 2.9 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.6 | 4.7 | GO:0007512 | adult heart development(GO:0007512) |
| 0.6 | 2.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.6 | 4.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.6 | 9.2 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.6 | 4.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 5.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.5 | 5.9 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.5 | 5.7 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.5 | 11.7 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.5 | 3.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.5 | 5.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.4 | 4.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.4 | 5.5 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.4 | 1.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 3.9 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 2.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 2.3 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.4 | 7.9 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.4 | 3.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 14.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.4 | 49.4 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.4 | 1.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 4.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 8.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 1.4 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.4 | 21.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.4 | 2.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.4 | 7.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.0 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.3 | 7.8 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.3 | 24.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.3 | 14.1 | GO:0006735 | NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 3.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.3 | 3.5 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.2 | 7.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 0.7 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 18.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 3.3 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 3.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 2.3 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 67.7 | GO:0007411 | axon guidance(GO:0007411) |
| 0.2 | 2.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.8 | GO:0051971 | negative regulation of transmission of nerve impulse(GO:0051970) positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.2 | 6.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 3.4 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 14.9 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.2 | 1.8 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.2 | 26.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 0.5 | GO:0097026 | myeloid dendritic cell chemotaxis(GO:0002408) dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 7.6 | GO:0007265 | Ras protein signal transduction(GO:0007265) |
| 0.2 | 1.1 | GO:0097396 | eosinophil differentiation(GO:0030222) response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.2 | 6.7 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.2 | 13.7 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.2 | 9.2 | GO:0003300 | cardiac muscle hypertrophy(GO:0003300) |
| 0.1 | 1.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 1.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 7.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 2.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 15.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 2.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.4 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 4.9 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 3.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 2.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.7 | GO:0042760 | positive regulation of fatty acid beta-oxidation(GO:0032000) very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 3.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 6.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 9.8 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.1 | 1.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 6.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 4.4 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.1 | 6.5 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.4 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 3.4 | GO:0007420 | brain development(GO:0007420) |
| 0.1 | 2.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 2.4 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.2 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 | 3.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 1.3 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 3.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.8 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 1.0 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 3.7 | GO:0005977 | glycogen metabolic process(GO:0005977) |
| 0.1 | 6.4 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 3.0 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 1.9 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 1.7 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 2.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.9 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.8 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.4 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.0 | 6.6 | GO:0046578 | regulation of Ras protein signal transduction(GO:0046578) |
| 0.0 | 32.5 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 7.0 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.5 | GO:0002517 | T cell tolerance induction(GO:0002517) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 7.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.0 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 1.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 2.1 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 1.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.3 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 4.8 | 115.8 | GO:0005861 | troponin complex(GO:0005861) |
| 4.5 | 26.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 3.9 | 46.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 3.5 | 69.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 3.0 | 109.0 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 3.0 | 12.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 2.5 | 27.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 2.3 | 13.9 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 2.3 | 9.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 2.2 | 8.8 | GO:0045160 | myosin I complex(GO:0045160) |
| 2.0 | 12.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 1.8 | 21.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 1.5 | 6.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 1.4 | 11.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.4 | 19.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.4 | 38.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 1.3 | 17.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.0 | 13.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.0 | 6.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 189.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.9 | 24.3 | GO:0036379 | myofilament(GO:0036379) |
| 0.8 | 56.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.6 | 101.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.6 | 13.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.6 | 1.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 2.3 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.6 | 23.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 13.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.6 | 19.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.5 | 16.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.4 | 1.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 3.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 8.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 3.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 46.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 4.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.2 | 15.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.2 | 8.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.2 | 3.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 14.3 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 7.8 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 2.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 14.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 8.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 9.2 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 3.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 10.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 6.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 17.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 19.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 3.8 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 4.6 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.1 | 32.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 1.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.1 | 4.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 23.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.1 | 0.5 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.2 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 3.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 5.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.8 | GO:0044439 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 6.4 | GO:0016021 | integral component of membrane(GO:0016021) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 27.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 5.1 | 15.3 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 4.9 | 24.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 4.9 | 19.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 4.5 | 63.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 4.4 | 13.3 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 4.2 | 25.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 3.9 | 89.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 3.9 | 15.6 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 3.7 | 11.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 3.2 | 9.5 | GO:0070538 | oleic acid binding(GO:0070538) |
| 3.1 | 12.5 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 3.1 | 46.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 3.0 | 12.0 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 3.0 | 15.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 2.8 | 11.4 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 2.6 | 13.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 2.5 | 4.9 | GO:0030172 | troponin C binding(GO:0030172) |
| 2.4 | 30.8 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.2 | 8.6 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.1 | 16.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 2.0 | 12.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 2.0 | 6.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 1.9 | 5.6 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 1.8 | 16.3 | GO:0031432 | titin binding(GO:0031432) |
| 1.8 | 19.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.8 | 10.6 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 1.6 | 22.7 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 1.5 | 51.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 1.5 | 10.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 1.4 | 8.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.4 | 19.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.4 | 5.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 1.3 | 70.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.1 | 9.2 | GO:0043559 | insulin binding(GO:0043559) |
| 1.1 | 4.5 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 1.1 | 3.2 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.9 | 4.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.9 | 13.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 5.0 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.8 | 7.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.7 | 52.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.7 | 3.7 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.7 | 2.9 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.7 | 2.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.7 | 19.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.7 | 6.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.6 | 1.9 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.6 | 3.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.6 | 4.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.6 | 2.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.6 | 2.3 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.6 | 2.8 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 7.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 8.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.5 | 2.6 | GO:0002046 | opsin binding(GO:0002046) |
| 0.5 | 10.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.5 | 2.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.4 | 7.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.4 | 3.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 7.5 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.4 | 2.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 2.3 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.4 | 4.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 8.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.4 | 1.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 11.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 4.6 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.4 | 3.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 6.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 5.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 2.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.3 | 3.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 6.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 5.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 5.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 9.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.3 | 7.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 1.0 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.3 | 1.8 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.2 | 3.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 17.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 1.6 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.2 | 10.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 4.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 17.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 2.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 4.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 0.8 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.2 | 4.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 15.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 4.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 6.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 2.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 57.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 59.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.5 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 93.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 9.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 8.2 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.5 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.1 | 3.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 3.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 1.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 6.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 9.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 2.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 12.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 8.6 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 5.9 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 4.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 9.6 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 26.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.9 | 35.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.7 | 98.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.7 | 23.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.6 | 6.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 109.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.4 | 58.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.4 | 5.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 17.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 11.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 7.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 9.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 10.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 5.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 5.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 8.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 4.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 2.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 3.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 3.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 236.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 1.3 | 18.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.9 | 9.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.9 | 19.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.8 | 7.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.7 | 13.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 17.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 35.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.5 | 16.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 7.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 9.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 21.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 3.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.4 | 5.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 8.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 12.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 17.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.3 | 3.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.3 | 10.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 16.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 14.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 5.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 7.8 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.3 | 5.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 11.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 2.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 12.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 4.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 2.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 6.4 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.2 | 6.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 5.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 4.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 4.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 5.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 7.6 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.1 | 3.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 2.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 4.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |