Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
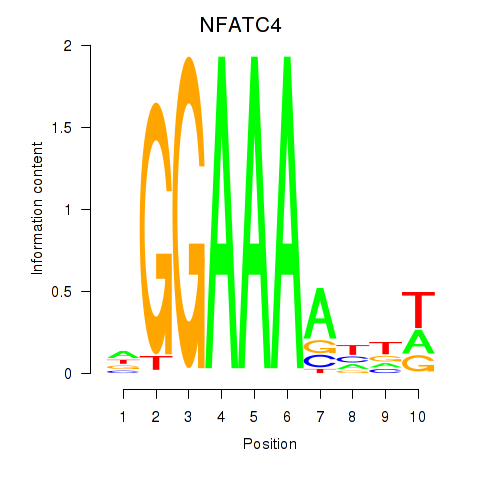
Results for NFATC4
Z-value: 0.50
Transcription factors associated with NFATC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFATC4
|
ENSG00000100968.9 | nuclear factor of activated T cells 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFATC4 | hg19_v2_chr14_+_24837226_24837547 | 0.32 | 9.9e-07 | Click! |
Activity profile of NFATC4 motif
Sorted Z-values of NFATC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_13835147 | 29.33 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr15_+_32933866 | 23.25 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr4_+_41258786 | 22.63 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr2_-_175712270 | 21.11 |
ENST00000295497.7
ENST00000444394.1 |
CHN1
|
chimerin 1 |
| chr8_-_22089845 | 19.04 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr14_+_29236269 | 18.02 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr3_-_98241760 | 17.80 |
ENST00000507874.1
ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1
|
claudin domain containing 1 |
| chr8_-_120685608 | 17.09 |
ENST00000427067.2
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_42811986 | 16.68 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr11_+_62475130 | 16.65 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr4_+_158142750 | 15.37 |
ENST00000505888.1
ENST00000449365.1 |
GRIA2
|
glutamate receptor, ionotropic, AMPA 2 |
| chr5_-_42812143 | 15.19 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_-_176733897 | 14.52 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr8_-_82359662 | 14.41 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr11_-_12030629 | 14.25 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr22_-_39548627 | 14.07 |
ENST00000216133.5
|
CBX7
|
chromobox homolog 7 |
| chr12_-_9268707 | 13.53 |
ENST00000318602.7
|
A2M
|
alpha-2-macroglobulin |
| chr6_-_152639479 | 13.38 |
ENST00000356820.4
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_163038565 | 13.23 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr14_-_74551096 | 13.11 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr1_-_57045228 | 12.43 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr1_+_10292308 | 12.16 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr12_+_12938541 | 11.57 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr19_-_39390440 | 10.84 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr5_+_140254884 | 10.83 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr11_-_62474803 | 10.74 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chrX_+_134166333 | 10.69 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr8_-_110703819 | 10.65 |
ENST00000532779.1
ENST00000534578.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr14_-_74551172 | 10.56 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chrX_-_49056635 | 10.52 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chrX_+_102631844 | 10.35 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr14_+_29234870 | 10.12 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr10_+_81838792 | 9.97 |
ENST00000372273.3
|
TMEM254
|
transmembrane protein 254 |
| chrX_-_43741594 | 9.89 |
ENST00000536181.1
ENST00000378069.4 |
MAOB
|
monoamine oxidase B |
| chr4_+_156587853 | 9.71 |
ENST00000506455.1
ENST00000511108.1 |
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr12_+_51318513 | 9.65 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr19_-_39390350 | 9.51 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr15_+_62853562 | 9.36 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr18_-_5540471 | 9.32 |
ENST00000581833.1
ENST00000544123.1 ENST00000342933.3 ENST00000400111.3 ENST00000585142.1 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr17_+_48610074 | 9.31 |
ENST00000503690.1
ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3
|
epsin 3 |
| chr8_+_85097110 | 9.20 |
ENST00000517638.1
ENST00000522647.1 |
RALYL
|
RALY RNA binding protein-like |
| chr6_+_123100620 | 9.11 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr19_+_30863271 | 9.06 |
ENST00000355537.3
|
ZNF536
|
zinc finger protein 536 |
| chr17_-_29624343 | 8.99 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr1_+_162602244 | 8.98 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr2_-_55277654 | 8.93 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr3_-_114035026 | 8.82 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr15_-_90892669 | 8.75 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr14_+_32546274 | 8.73 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr17_+_53343577 | 8.72 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr2_+_191208196 | 8.59 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr2_-_55277692 | 8.56 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr16_+_2039946 | 8.53 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chr8_-_19459993 | 8.39 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr1_+_164528866 | 8.03 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_+_76311791 | 7.98 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr4_+_156588249 | 7.77 |
ENST00000393832.3
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr8_-_18541603 | 7.74 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr4_+_156588115 | 7.58 |
ENST00000455639.2
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr6_-_167275991 | 7.46 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr14_+_85996471 | 7.42 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr4_+_113970772 | 7.31 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr12_-_54582655 | 7.28 |
ENST00000504338.1
ENST00000514685.1 ENST00000504797.1 ENST00000513838.1 ENST00000505128.1 ENST00000337581.3 ENST00000503306.1 ENST00000243112.5 ENST00000514196.1 ENST00000506169.1 ENST00000507904.1 ENST00000508394.2 |
SMUG1
|
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| chr6_-_134495992 | 7.09 |
ENST00000475719.2
ENST00000367857.5 ENST00000237305.7 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr7_+_20686946 | 7.07 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr19_+_18794470 | 7.01 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr6_-_154677900 | 6.75 |
ENST00000265198.4
ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr8_+_28747884 | 6.72 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr4_+_156587979 | 6.59 |
ENST00000511507.1
|
GUCY1A3
|
guanylate cyclase 1, soluble, alpha 3 |
| chr16_+_57769635 | 6.49 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr4_-_153303658 | 6.25 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr4_-_166034029 | 6.14 |
ENST00000306480.6
|
TMEM192
|
transmembrane protein 192 |
| chr2_+_27237615 | 6.05 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr12_+_101988774 | 6.00 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr12_+_41086297 | 5.98 |
ENST00000551295.2
|
CNTN1
|
contactin 1 |
| chr2_+_173600514 | 5.79 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_+_101988627 | 5.78 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr19_+_52772832 | 5.67 |
ENST00000593703.1
ENST00000601711.1 ENST00000599581.1 |
ZNF766
|
zinc finger protein 766 |
| chr9_+_706842 | 5.63 |
ENST00000382293.3
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr6_-_167276033 | 5.57 |
ENST00000503859.1
ENST00000506565.1 |
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr22_-_36236623 | 5.50 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr7_-_152373216 | 5.43 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr3_-_33686743 | 5.43 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr14_+_65171315 | 5.29 |
ENST00000394691.1
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr14_+_94577074 | 5.15 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr1_-_113162040 | 5.15 |
ENST00000358039.4
ENST00000369668.2 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr14_+_100531615 | 5.14 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr14_+_22984601 | 5.14 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr14_-_30396948 | 5.10 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr3_-_15106747 | 5.06 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chr14_+_65171099 | 5.00 |
ENST00000247226.7
|
PLEKHG3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_+_173600565 | 4.97 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr6_+_158733692 | 4.94 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr2_-_55276320 | 4.91 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr7_-_14942283 | 4.82 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr12_-_56106060 | 4.80 |
ENST00000452168.2
|
ITGA7
|
integrin, alpha 7 |
| chr2_-_152830479 | 4.77 |
ENST00000360283.6
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr15_-_77712477 | 4.76 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr9_+_104161123 | 4.71 |
ENST00000374861.3
ENST00000339664.2 ENST00000259395.4 |
ZNF189
|
zinc finger protein 189 |
| chr19_+_52772821 | 4.71 |
ENST00000439461.1
|
ZNF766
|
zinc finger protein 766 |
| chr2_+_171673072 | 4.61 |
ENST00000358196.3
ENST00000375272.1 |
GAD1
|
glutamate decarboxylase 1 (brain, 67kDa) |
| chr15_+_43985084 | 4.57 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr22_-_36220420 | 4.36 |
ENST00000473487.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr18_+_5238549 | 4.25 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr10_+_81838411 | 4.23 |
ENST00000372281.3
ENST00000372277.3 ENST00000372275.1 ENST00000372274.1 |
TMEM254
|
transmembrane protein 254 |
| chr1_+_174846570 | 4.22 |
ENST00000392064.2
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr12_+_26348246 | 4.17 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr1_+_50569575 | 4.12 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr17_-_58603568 | 4.05 |
ENST00000083182.3
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr18_-_64271363 | 4.02 |
ENST00000262150.2
|
CDH19
|
cadherin 19, type 2 |
| chr3_-_18480260 | 4.02 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr19_-_14887568 | 4.01 |
ENST00000596991.2
ENST00000594294.1 ENST00000594076.1 ENST00000595839.1 ENST00000392965.3 ENST00000601345.1 |
EMR2
|
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr5_-_88179017 | 3.98 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr3_+_118905564 | 3.94 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr19_-_15087839 | 3.88 |
ENST00000600144.1
|
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr15_+_43885252 | 3.85 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr4_+_41540160 | 3.84 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_114790179 | 3.78 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr4_-_46391805 | 3.73 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr7_-_135412925 | 3.71 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chrX_-_102941596 | 3.69 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr14_+_75536335 | 3.66 |
ENST00000554763.1
ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr11_+_19799327 | 3.65 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr12_-_30887948 | 3.63 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chr3_-_46069223 | 3.61 |
ENST00000309285.3
|
XCR1
|
chemokine (C motif) receptor 1 |
| chr4_-_46391931 | 3.51 |
ENST00000381620.4
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr5_-_88178964 | 3.49 |
ENST00000513252.1
ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C
|
myocyte enhancer factor 2C |
| chr14_+_85996507 | 3.39 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr12_-_123450986 | 3.38 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr3_-_73673991 | 3.34 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr6_-_39902185 | 3.31 |
ENST00000373195.3
ENST00000308559.7 ENST00000373188.2 |
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr15_+_63354769 | 3.26 |
ENST00000558910.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_-_49137790 | 3.26 |
ENST00000599385.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr14_+_96858454 | 3.24 |
ENST00000555570.1
|
AK7
|
adenylate kinase 7 |
| chr7_+_18535786 | 3.23 |
ENST00000406072.1
|
HDAC9
|
histone deacetylase 9 |
| chr5_+_149109825 | 3.21 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr12_+_27396901 | 3.21 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr11_-_61735029 | 3.19 |
ENST00000526640.1
|
FTH1
|
ferritin, heavy polypeptide 1 |
| chr5_+_15500280 | 3.18 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr19_-_17488143 | 3.13 |
ENST00000599426.1
ENST00000252590.4 |
PLVAP
|
plasmalemma vesicle associated protein |
| chr14_+_75536280 | 3.12 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr14_+_57857262 | 3.12 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr9_+_92219919 | 3.10 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr20_-_8000426 | 3.09 |
ENST00000527925.1
ENST00000246024.2 |
TMX4
|
thioredoxin-related transmembrane protein 4 |
| chr9_-_13165457 | 3.09 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr8_+_98900132 | 3.06 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr9_+_91933407 | 3.06 |
ENST00000375807.3
ENST00000339901.4 |
SECISBP2
|
SECIS binding protein 2 |
| chr17_-_4607335 | 3.03 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr10_+_123872483 | 3.02 |
ENST00000369001.1
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr1_+_36789335 | 3.01 |
ENST00000373137.2
|
RP11-268J15.5
|
RP11-268J15.5 |
| chrX_+_76709648 | 2.98 |
ENST00000439435.1
|
FGF16
|
fibroblast growth factor 16 |
| chr8_+_94929969 | 2.94 |
ENST00000517764.1
|
PDP1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_-_44657017 | 2.93 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr5_-_43412418 | 2.93 |
ENST00000537013.1
ENST00000361115.4 |
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr6_-_131384373 | 2.92 |
ENST00000392427.3
ENST00000525271.1 ENST00000527411.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chrX_+_10124977 | 2.88 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_-_131384347 | 2.88 |
ENST00000530481.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr19_-_49137762 | 2.88 |
ENST00000593500.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr9_-_115818951 | 2.86 |
ENST00000553380.1
ENST00000374227.3 |
ZFP37
|
ZFP37 zinc finger protein |
| chr19_-_53606604 | 2.86 |
ENST00000599056.1
ENST00000599247.1 ENST00000355147.5 ENST00000429604.1 ENST00000418871.1 ENST00000599637.1 |
ZNF160
|
zinc finger protein 160 |
| chr12_-_90049878 | 2.84 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr19_-_1174226 | 2.83 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr12_-_90049828 | 2.78 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr20_+_57875758 | 2.75 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr17_+_5389605 | 2.74 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr6_-_131384412 | 2.73 |
ENST00000445890.2
ENST00000368128.2 ENST00000337057.3 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr19_+_16296191 | 2.72 |
ENST00000589852.1
ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32, member A |
| chr5_+_131993856 | 2.71 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr17_-_18430160 | 2.70 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chrX_+_22056165 | 2.69 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr12_-_49582978 | 2.67 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_-_89744314 | 2.64 |
ENST00000508369.1
|
FAM13A
|
family with sequence similarity 13, member A |
| chr12_+_109577202 | 2.61 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr1_-_113161730 | 2.61 |
ENST00000544629.1
ENST00000543570.1 ENST00000360743.4 ENST00000490067.1 ENST00000343210.7 ENST00000369666.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr12_+_26348429 | 2.59 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr6_-_39902160 | 2.56 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chr17_+_37793378 | 2.54 |
ENST00000544210.2
ENST00000581894.1 ENST00000394250.4 ENST00000579479.1 ENST00000577248.1 ENST00000580611.1 |
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr2_-_177502254 | 2.50 |
ENST00000339037.3
|
AC017048.3
|
long intergenic non-protein coding RNA 1116 |
| chr3_+_186560476 | 2.47 |
ENST00000320741.2
ENST00000444204.2 |
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560462 | 2.46 |
ENST00000412955.2
|
ADIPOQ
|
adiponectin, C1Q and collagen domain containing |
| chr7_+_138943265 | 2.46 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr6_+_87865262 | 2.42 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr17_+_45331184 | 2.42 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr14_-_24711470 | 2.42 |
ENST00000559969.1
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr12_-_95467267 | 2.41 |
ENST00000330677.7
|
NR2C1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr2_+_220306745 | 2.39 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr9_-_80263220 | 2.33 |
ENST00000341700.6
|
GNA14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr13_-_33859819 | 2.33 |
ENST00000336934.5
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr6_-_33290580 | 2.28 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chrX_-_77041685 | 2.25 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chr3_+_155838337 | 2.19 |
ENST00000490337.1
ENST00000389636.5 |
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr4_-_89744365 | 2.17 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr14_-_30396802 | 2.16 |
ENST00000415220.2
|
PRKD1
|
protein kinase D1 |
| chr3_-_194090460 | 2.16 |
ENST00000428839.1
ENST00000347624.3 |
LRRC15
|
leucine rich repeat containing 15 |
| chr15_-_86338134 | 2.15 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr6_-_146057144 | 2.15 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr13_+_97874574 | 2.13 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr7_+_5919458 | 2.11 |
ENST00000416608.1
|
OCM
|
oncomodulin |
| chrX_-_133119895 | 2.11 |
ENST00000370818.3
|
GPC3
|
glypican 3 |
| chr8_+_17434689 | 2.10 |
ENST00000398074.3
|
PDGFRL
|
platelet-derived growth factor receptor-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFATC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 6.8 | 20.4 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 5.9 | 23.7 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 5.3 | 31.7 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 4.1 | 12.2 | GO:1904647 | response to rotenone(GO:1904647) |
| 3.6 | 14.2 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 2.9 | 29.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.5 | 22.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 2.4 | 7.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.3 | 7.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.1 | 8.4 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 2.0 | 6.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.9 | 5.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 1.9 | 7.5 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 1.8 | 7.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 1.8 | 10.8 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 1.8 | 7.1 | GO:0048749 | compound eye development(GO:0048749) |
| 1.7 | 10.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.6 | 6.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 1.3 | 13.4 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.3 | 9.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 1.3 | 6.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 1.2 | 8.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 1.2 | 14.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 1.1 | 5.6 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 1.0 | 13.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 1.0 | 7.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 1.0 | 12.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 1.0 | 15.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.0 | 7.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 1.0 | 11.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.9 | 3.7 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.9 | 3.7 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.9 | 5.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.9 | 2.7 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.8 | 1.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.8 | 3.9 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.7 | 1.5 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.7 | 5.9 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.7 | 5.5 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.7 | 2.7 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.7 | 3.3 | GO:0051136 | extrathymic T cell differentiation(GO:0033078) regulation of NK T cell differentiation(GO:0051136) |
| 0.7 | 2.6 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.6 | 9.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 8.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.6 | 2.9 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.6 | 9.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.6 | 5.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.6 | 2.3 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.6 | 2.3 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.6 | 3.4 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.6 | 7.3 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.5 | 7.6 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.5 | 2.7 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.5 | 2.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.5 | 3.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.5 | 18.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.5 | 3.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.5 | 8.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 1.0 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.5 | 19.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.5 | 2.8 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.5 | 4.2 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.5 | 2.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.5 | 3.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.4 | 6.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 10.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.4 | 3.0 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.4 | 1.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.4 | 5.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.4 | 4.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 3.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 | 2.4 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.4 | 7.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.4 | 3.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.4 | 1.5 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.4 | 1.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.4 | 12.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 3.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 3.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 2.0 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.3 | 13.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 9.3 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.3 | 16.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.3 | 15.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 2.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.3 | 1.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.3 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.3 | 1.6 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.3 | 2.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.3 | 2.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 1.4 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.2 | 3.8 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.2 | 2.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 1.8 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.9 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 4.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.2 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 1.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 4.0 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.2 | 3.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 0.7 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 1.5 | GO:1902949 | positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.2 | 1.3 | GO:0072513 | semicircular canal morphogenesis(GO:0048752) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 3.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.2 | 4.2 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 0.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.2 | 5.1 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.2 | 10.8 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 9.0 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.1 | 6.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 3.2 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.1 | 8.5 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.1 | 0.6 | GO:2000110 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.9 | GO:0014827 | intestine smooth muscle contraction(GO:0014827) |
| 0.1 | 0.8 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 3.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 6.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 2.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 8.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 2.1 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.1 | 0.8 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 6.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.4 | GO:0021938 | ventral midline development(GO:0007418) cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.1 | 4.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 8.6 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 4.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 17.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.6 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 1.5 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.1 | 0.9 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 3.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 5.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 3.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.1 | 2.9 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.1 | 0.9 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 2.0 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 3.1 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 3.1 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.1 | 4.1 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.9 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 4.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 3.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 4.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 2.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 4.6 | GO:0030879 | mammary gland development(GO:0030879) |
| 0.0 | 1.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 1.3 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 2.3 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.8 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 1.2 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 1.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 1.4 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 7.8 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 1.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 1.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 5.5 | GO:0007420 | brain development(GO:0007420) |
| 0.0 | 0.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.4 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 2.3 | 20.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.0 | 5.9 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 1.6 | 4.8 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 1.3 | 31.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.1 | 22.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 1.0 | 17.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.0 | 13.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.9 | 5.4 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.9 | 6.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.9 | 12.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.8 | 10.6 | GO:0097433 | dense body(GO:0097433) |
| 0.8 | 2.4 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.8 | 3.2 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.8 | 3.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.7 | 34.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.6 | 9.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.6 | 8.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 2.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.5 | 5.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.5 | 11.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.4 | 16.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.4 | 2.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 5.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 3.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.3 | 3.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.3 | 3.7 | GO:0060077 | inhibitory synapse(GO:0060077) clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.3 | 14.9 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 5.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 7.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 14.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 1.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.7 | GO:0043159 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.2 | 8.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 3.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 6.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 3.3 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 5.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 2.3 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 3.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 12.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 1.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 9.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 3.0 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 3.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 4.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.2 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.1 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 5.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 8.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 11.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 5.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 5.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 6.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 9.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 8.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 5.7 | GO:0044450 | microtubule organizing center part(GO:0044450) |
| 0.0 | 10.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 7.1 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 9.1 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.9 | GO:0019867 | outer membrane(GO:0019867) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.9 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 5.7 | 22.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 4.5 | 13.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 4.3 | 17.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 2.8 | 8.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 1.8 | 7.3 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 1.8 | 9.0 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.4 | 5.5 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.3 | 15.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.3 | 31.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 1.1 | 9.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.1 | 3.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 1.1 | 6.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.1 | 4.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 1.0 | 8.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.0 | 7.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 1.0 | 16.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.9 | 23.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.9 | 8.6 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.9 | 8.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.8 | 3.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.8 | 11.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.8 | 6.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.8 | 3.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.8 | 2.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.7 | 7.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.7 | 2.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.7 | 6.7 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.7 | 18.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.7 | 2.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.6 | 1.9 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.6 | 1.8 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.6 | 29.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.6 | 4.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 2.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 3.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.5 | 3.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.5 | 2.1 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.5 | 7.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 6.7 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 3.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.5 | 2.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 5.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.5 | 7.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 3.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.5 | 3.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 20.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.4 | 13.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 2.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.4 | 1.5 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.4 | 3.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 5.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 10.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 4.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 2.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.3 | 15.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 7.6 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.3 | 2.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 7.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 5.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 7.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 4.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 3.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 0.8 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 5.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 12.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.2 | 3.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 4.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 10.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 0.9 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.2 | 2.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.2 | 5.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 7.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 2.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 2.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 3.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 4.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.5 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 11.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 17.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 5.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 4.1 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 4.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 2.1 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 14.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 7.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 9.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.9 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 2.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 1.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 3.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 2.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.2 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 1.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 4.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 6.9 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 7.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 5.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 6.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 6.6 | GO:0008289 | lipid binding(GO:0008289) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 20.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 32.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 6.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 41.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 15.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.3 | 21.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 13.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 5.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.3 | 2.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 5.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 2.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 8.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 10.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.2 | 7.1 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 6.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 8.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 8.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 4.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 3.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 6.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 1.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.3 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 2.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.0 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 5.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 3.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 20.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.7 | 16.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.7 | 13.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 15.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.6 | 30.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.5 | 17.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.4 | 7.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.4 | 5.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 2.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 38.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.3 | 6.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 11.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 10.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 7.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 9.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 6.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 7.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 2.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 12.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 4.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 20.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 10.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 14.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 6.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 5.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 7.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.8 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 4.8 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 3.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.1 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.1 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.0 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 4.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 4.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |