Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
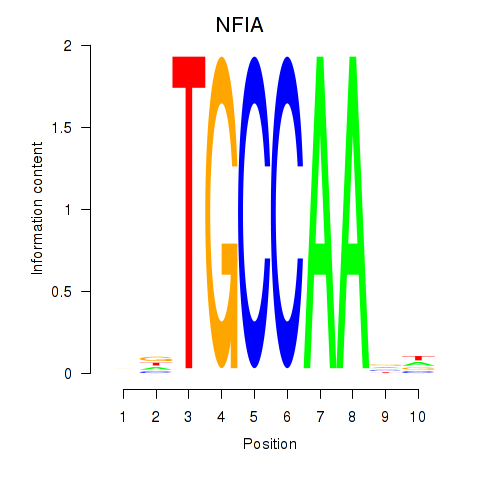
Results for NFIA
Z-value: 1.01
Transcription factors associated with NFIA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFIA
|
ENSG00000162599.11 | nuclear factor I A |
Activity profile of NFIA motif
Sorted Z-values of NFIA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_29171689 | 17.84 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr16_+_58283814 | 13.44 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr12_-_15038779 | 12.64 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr4_-_57547870 | 12.32 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr14_+_73704201 | 11.39 |
ENST00000340738.5
ENST00000427855.1 ENST00000381166.3 |
PAPLN
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr4_-_57547454 | 10.91 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr14_+_23299088 | 9.80 |
ENST00000355151.5
ENST00000397496.3 ENST00000555345.1 ENST00000432849.3 ENST00000553711.1 ENST00000556465.1 ENST00000397505.2 ENST00000557221.1 ENST00000311892.6 ENST00000556840.1 ENST00000555536.1 |
MRPL52
|
mitochondrial ribosomal protein L52 |
| chr14_+_75746340 | 9.48 |
ENST00000555686.1
ENST00000555672.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr8_-_22089533 | 9.40 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr9_+_17135016 | 8.85 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr14_+_75746781 | 8.68 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr9_+_17134980 | 8.00 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr3_+_109128837 | 7.66 |
ENST00000497996.1
|
RP11-702L6.4
|
RP11-702L6.4 |
| chr19_-_46296011 | 7.64 |
ENST00000377735.3
ENST00000270223.6 |
DMWD
|
dystrophia myotonica, WD repeat containing |
| chr3_+_45067659 | 7.41 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr14_-_60337684 | 7.33 |
ENST00000267484.5
|
RTN1
|
reticulon 1 |
| chr22_-_38699003 | 7.30 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr4_-_176733897 | 7.16 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr11_-_59383617 | 7.15 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr18_+_61144160 | 7.15 |
ENST00000489441.1
ENST00000424602.1 |
SERPINB5
|
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr4_+_74269956 | 6.76 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr11_-_5276008 | 6.65 |
ENST00000336906.4
|
HBG2
|
hemoglobin, gamma G |
| chr9_+_130911770 | 6.50 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr5_+_40909354 | 6.43 |
ENST00000313164.9
|
C7
|
complement component 7 |
| chr17_-_1733114 | 6.36 |
ENST00000305513.7
|
SMYD4
|
SET and MYND domain containing 4 |
| chr1_-_203320617 | 6.35 |
ENST00000354955.4
|
FMOD
|
fibromodulin |
| chr9_-_93405352 | 6.29 |
ENST00000375765.3
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2 |
| chrX_+_103031758 | 6.24 |
ENST00000303958.2
ENST00000361621.2 |
PLP1
|
proteolipid protein 1 |
| chr5_-_34043310 | 6.21 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr9_+_130911723 | 6.18 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr10_+_5005598 | 6.03 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr17_-_4167142 | 5.97 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr4_-_87028478 | 5.95 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr16_-_55867146 | 5.83 |
ENST00000422046.2
|
CES1
|
carboxylesterase 1 |
| chr15_+_81475047 | 5.80 |
ENST00000559388.1
|
IL16
|
interleukin 16 |
| chr19_+_49258775 | 5.78 |
ENST00000593756.1
|
FGF21
|
fibroblast growth factor 21 |
| chrX_-_13835147 | 5.77 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr9_+_117085336 | 5.76 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr19_-_40324767 | 5.75 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr5_+_121647386 | 5.71 |
ENST00000542191.1
ENST00000506272.1 ENST00000508681.1 ENST00000509154.2 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr16_+_6533729 | 5.71 |
ENST00000551752.1
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr12_-_10251576 | 5.69 |
ENST00000315330.4
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr11_-_5271122 | 5.60 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr3_+_101498074 | 5.57 |
ENST00000273347.5
ENST00000474165.1 |
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr3_-_52860850 | 5.54 |
ENST00000441637.2
|
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr7_-_8302207 | 5.49 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr2_+_30569506 | 5.43 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr7_-_8301869 | 5.37 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr16_-_55866997 | 5.36 |
ENST00000360526.3
ENST00000361503.4 |
CES1
|
carboxylesterase 1 |
| chr1_-_226926864 | 5.34 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr17_-_8066250 | 5.34 |
ENST00000488857.1
ENST00000481878.1 ENST00000316509.6 ENST00000498285.1 |
VAMP2
RP11-599B13.6
|
vesicle-associated membrane protein 2 (synaptobrevin 2) Uncharacterized protein |
| chr14_+_22964877 | 5.31 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr1_+_22962948 | 5.29 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr2_+_241938255 | 5.28 |
ENST00000401884.1
ENST00000405547.3 ENST00000310397.8 ENST00000342631.6 |
SNED1
|
sushi, nidogen and EGF-like domains 1 |
| chr12_+_113229737 | 5.20 |
ENST00000551052.1
ENST00000415485.3 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr1_+_118148556 | 5.18 |
ENST00000369448.3
|
FAM46C
|
family with sequence similarity 46, member C |
| chr7_-_8302164 | 5.17 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr6_-_49681235 | 5.16 |
ENST00000339139.4
|
CRISP2
|
cysteine-rich secretory protein 2 |
| chr17_-_10452929 | 5.16 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chr7_+_153749732 | 5.10 |
ENST00000377770.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr9_-_99064429 | 5.08 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr12_-_10251603 | 5.04 |
ENST00000457018.2
|
CLEC1A
|
C-type lectin domain family 1, member A |
| chr6_+_26156551 | 5.02 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr3_-_114343768 | 5.02 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_-_208989225 | 5.00 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chrX_+_103031421 | 4.90 |
ENST00000433491.1
ENST00000418604.1 ENST00000443502.1 |
PLP1
|
proteolipid protein 1 |
| chr3_-_194072019 | 4.88 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr12_-_53207842 | 4.88 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr2_+_189839046 | 4.87 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr12_+_113229543 | 4.86 |
ENST00000447659.2
|
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr12_-_99038732 | 4.86 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr17_+_34430980 | 4.83 |
ENST00000250151.4
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr2_-_176866978 | 4.83 |
ENST00000392540.2
ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715
|
KIAA1715 |
| chr2_+_17721230 | 4.81 |
ENST00000457525.1
|
VSNL1
|
visinin-like 1 |
| chr3_+_111718173 | 4.75 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr3_+_4535155 | 4.74 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_+_126225789 | 4.71 |
ENST00000530591.1
ENST00000534083.1 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr12_-_13248562 | 4.69 |
ENST00000457134.2
ENST00000537302.1 |
GSG1
|
germ cell associated 1 |
| chr18_-_5396271 | 4.64 |
ENST00000579951.1
|
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr6_+_150920999 | 4.62 |
ENST00000367328.1
ENST00000367326.1 |
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr16_+_67840986 | 4.58 |
ENST00000561639.1
ENST00000567852.1 ENST00000565148.1 ENST00000388833.3 ENST00000561654.1 ENST00000431934.2 |
TSNAXIP1
|
translin-associated factor X interacting protein 1 |
| chr12_-_13248705 | 4.57 |
ENST00000396310.2
|
GSG1
|
germ cell associated 1 |
| chrX_+_122993827 | 4.52 |
ENST00000371199.3
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr17_-_10421853 | 4.48 |
ENST00000226207.5
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult |
| chrX_+_69642881 | 4.45 |
ENST00000453994.2
ENST00000536730.1 ENST00000538649.1 ENST00000374382.3 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr9_+_137967366 | 4.44 |
ENST00000252854.4
|
OLFM1
|
olfactomedin 1 |
| chr14_-_76447494 | 4.42 |
ENST00000238682.3
|
TGFB3
|
transforming growth factor, beta 3 |
| chr9_+_37667978 | 4.41 |
ENST00000539465.1
|
FRMPD1
|
FERM and PDZ domain containing 1 |
| chr13_-_36429763 | 4.40 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr4_+_113739244 | 4.37 |
ENST00000503271.1
ENST00000503423.1 ENST00000506722.1 |
ANK2
|
ankyrin 2, neuronal |
| chr18_+_77155856 | 4.30 |
ENST00000253506.5
ENST00000591814.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr9_+_137967268 | 4.29 |
ENST00000371799.4
ENST00000277415.11 |
OLFM1
|
olfactomedin 1 |
| chr1_+_198608146 | 4.29 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chrX_-_13835461 | 4.26 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr19_-_40440533 | 4.23 |
ENST00000221347.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chr4_+_71263599 | 4.20 |
ENST00000399575.2
|
PROL1
|
proline rich, lacrimal 1 |
| chr10_-_21463116 | 4.18 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chrX_+_122993657 | 4.13 |
ENST00000434753.3
ENST00000430625.1 |
XIAP
|
X-linked inhibitor of apoptosis |
| chr1_+_66797687 | 4.12 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_-_111150048 | 4.11 |
ENST00000485317.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr18_+_61575200 | 4.10 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr10_-_100995540 | 4.05 |
ENST00000370546.1
ENST00000404542.1 |
HPSE2
|
heparanase 2 |
| chr12_+_69864129 | 4.04 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr8_+_38585704 | 4.02 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr1_+_180601139 | 3.99 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr5_+_176853702 | 3.98 |
ENST00000507633.1
ENST00000393576.3 ENST00000355958.5 ENST00000528793.1 ENST00000512684.1 |
GRK6
|
G protein-coupled receptor kinase 6 |
| chr3_+_4535025 | 3.98 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr11_+_63870660 | 3.88 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr19_-_12997995 | 3.86 |
ENST00000264834.4
|
KLF1
|
Kruppel-like factor 1 (erythroid) |
| chr19_-_40324255 | 3.84 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr10_-_100995603 | 3.84 |
ENST00000370552.3
ENST00000370549.1 |
HPSE2
|
heparanase 2 |
| chr14_-_75079026 | 3.82 |
ENST00000261978.4
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr19_+_36249057 | 3.80 |
ENST00000301165.5
ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55
|
chromosome 19 open reading frame 55 |
| chr11_+_64004888 | 3.80 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr12_-_13248598 | 3.75 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chrX_-_135849484 | 3.70 |
ENST00000370620.1
ENST00000535227.1 |
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr17_+_61699766 | 3.64 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr17_+_72270380 | 3.63 |
ENST00000582036.1
ENST00000307504.5 |
DNAI2
|
dynein, axonemal, intermediate chain 2 |
| chr11_-_128392085 | 3.63 |
ENST00000526145.2
ENST00000531611.1 ENST00000319397.6 ENST00000345075.4 ENST00000535549.1 |
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr12_+_122064398 | 3.62 |
ENST00000330079.7
|
ORAI1
|
ORAI calcium release-activated calcium modulator 1 |
| chrX_+_37639264 | 3.61 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr7_+_94537542 | 3.56 |
ENST00000433881.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr13_+_23755099 | 3.56 |
ENST00000537476.1
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr8_-_21988558 | 3.53 |
ENST00000312841.8
|
HR
|
hair growth associated |
| chr12_-_13248732 | 3.52 |
ENST00000396302.3
|
GSG1
|
germ cell associated 1 |
| chr16_-_79634595 | 3.48 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr4_+_3076388 | 3.46 |
ENST00000355072.5
|
HTT
|
huntingtin |
| chr2_+_33701286 | 3.45 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr5_+_176853669 | 3.43 |
ENST00000355472.5
|
GRK6
|
G protein-coupled receptor kinase 6 |
| chrX_+_37639302 | 3.43 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr16_+_21312170 | 3.43 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr1_-_120311517 | 3.42 |
ENST00000369406.3
ENST00000544913.2 |
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) |
| chr5_-_55529115 | 3.42 |
ENST00000513241.2
ENST00000341048.4 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr11_+_47291193 | 3.39 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr2_-_158345462 | 3.39 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr17_+_29664830 | 3.37 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr3_+_121613265 | 3.36 |
ENST00000295605.2
|
SLC15A2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chrX_-_69479654 | 3.35 |
ENST00000374519.2
|
P2RY4
|
pyrimidinergic receptor P2Y, G-protein coupled, 4 |
| chr12_+_56324756 | 3.26 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr9_-_104198042 | 3.26 |
ENST00000374855.4
|
ALDOB
|
aldolase B, fructose-bisphosphate |
| chr15_+_59499031 | 3.25 |
ENST00000307144.4
|
LDHAL6B
|
lactate dehydrogenase A-like 6B |
| chr11_-_35547151 | 3.21 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr7_-_99381884 | 3.20 |
ENST00000336411.2
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr3_+_111260954 | 3.19 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr10_-_82049424 | 3.19 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chrX_+_122994112 | 3.14 |
ENST00000355640.3
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr8_+_124194752 | 3.12 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr12_+_8995832 | 3.09 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr7_-_99332719 | 3.06 |
ENST00000336374.2
|
CYP3A7
|
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr3_+_111260856 | 3.04 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr13_-_29069232 | 3.04 |
ENST00000282397.4
ENST00000541932.1 ENST00000539099.1 |
FLT1
|
fms-related tyrosine kinase 1 |
| chr14_-_98444386 | 3.01 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr1_-_21978312 | 3.00 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr13_+_23755054 | 2.97 |
ENST00000218867.3
|
SGCG
|
sarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) |
| chr7_-_97619508 | 2.96 |
ENST00000257627.4
|
OCM2
|
oncomodulin 2 |
| chr19_+_12949251 | 2.96 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr8_+_133931648 | 2.96 |
ENST00000519178.1
ENST00000542445.1 |
TG
|
thyroglobulin |
| chr7_+_147830776 | 2.95 |
ENST00000538075.1
|
CNTNAP2
|
contactin associated protein-like 2 |
| chr5_+_36608422 | 2.92 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_-_39769005 | 2.90 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr12_+_58138664 | 2.90 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr1_-_111148241 | 2.87 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr16_-_15474904 | 2.87 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr1_-_236046872 | 2.86 |
ENST00000536965.1
|
LYST
|
lysosomal trafficking regulator |
| chr2_+_87565634 | 2.85 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr22_+_41763274 | 2.83 |
ENST00000406644.3
|
TEF
|
thyrotrophic embryonic factor |
| chr7_-_135433460 | 2.83 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr1_-_201081579 | 2.82 |
ENST00000367338.3
ENST00000362061.3 |
CACNA1S
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr6_-_32784687 | 2.81 |
ENST00000447394.1
ENST00000438763.2 |
HLA-DOB
|
major histocompatibility complex, class II, DO beta |
| chr4_+_113970772 | 2.81 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr2_-_90538397 | 2.79 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr12_+_56114151 | 2.79 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_+_49259325 | 2.77 |
ENST00000222157.3
|
FGF21
|
fibroblast growth factor 21 |
| chr9_+_71944241 | 2.76 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr20_+_42136308 | 2.74 |
ENST00000434666.1
ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr3_+_148508845 | 2.74 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chrX_+_22050546 | 2.73 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr15_+_58702742 | 2.73 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr5_+_152870106 | 2.72 |
ENST00000285900.5
|
GRIA1
|
glutamate receptor, ionotropic, AMPA 1 |
| chr17_-_8093471 | 2.72 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr2_-_160761179 | 2.71 |
ENST00000554112.1
ENST00000553424.1 ENST00000263636.4 ENST00000504764.1 ENST00000505052.1 |
LY75
LY75-CD302
|
lymphocyte antigen 75 LY75-CD302 readthrough |
| chr1_+_209878182 | 2.70 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr4_+_71226468 | 2.69 |
ENST00000226460.4
|
SMR3A
|
submaxillary gland androgen regulated protein 3A |
| chr5_+_161494521 | 2.66 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr12_+_100897130 | 2.66 |
ENST00000551379.1
ENST00000188403.7 ENST00000551184.1 |
NR1H4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr1_-_175712665 | 2.65 |
ENST00000263525.2
|
TNR
|
tenascin R |
| chr5_+_67586465 | 2.65 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr9_+_36169380 | 2.63 |
ENST00000335119.2
|
CCIN
|
calicin |
| chr9_+_27109133 | 2.62 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr12_+_58138800 | 2.62 |
ENST00000547992.1
ENST00000552816.1 ENST00000547472.1 |
TSPAN31
|
tetraspanin 31 |
| chr12_+_56114189 | 2.62 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chrX_-_85302531 | 2.59 |
ENST00000537751.1
ENST00000358786.4 ENST00000357749.2 |
CHM
|
choroideremia (Rab escort protein 1) |
| chr2_+_3705785 | 2.58 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr5_-_64777733 | 2.54 |
ENST00000381055.3
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr2_+_68592305 | 2.53 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr16_+_30383613 | 2.52 |
ENST00000568749.1
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_-_211036051 | 2.51 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr6_-_30043539 | 2.46 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr5_+_140868717 | 2.46 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr11_-_62389621 | 2.44 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NFIA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.8 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 2.5 | 12.7 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.4 | 7.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 2.3 | 18.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 2.3 | 6.8 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 2.1 | 8.6 | GO:1904640 | response to methionine(GO:1904640) |
| 2.1 | 6.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 2.1 | 6.2 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 1.7 | 7.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 1.7 | 8.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 1.6 | 4.9 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 1.5 | 19.6 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 1.5 | 4.5 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.4 | 7.0 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.3 | 4.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 1.2 | 3.5 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 1.1 | 3.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 1.1 | 6.7 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 1.1 | 11.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 1.1 | 8.9 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 1.1 | 4.4 | GO:1905071 | ossification involved in bone remodeling(GO:0043932) frontal suture morphogenesis(GO:0060364) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 1.1 | 5.5 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 1.1 | 3.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.1 | 4.3 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 1.0 | 23.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 1.0 | 10.0 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.0 | 3.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 1.0 | 3.0 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 1.0 | 8.7 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 1.0 | 2.9 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.9 | 5.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.9 | 2.8 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.9 | 16.8 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.9 | 2.8 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.9 | 3.6 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.9 | 5.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.9 | 2.6 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.9 | 4.3 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.9 | 11.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.8 | 5.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 12.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.8 | 3.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.8 | 2.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.8 | 5.3 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.8 | 5.3 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.7 | 5.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 5.8 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.7 | 2.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 0.7 | 4.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.7 | 1.4 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.7 | 2.7 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.7 | 4.6 | GO:0071205 | protein localization to paranode region of axon(GO:0002175) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.6 | 3.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.6 | 5.0 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.6 | 3.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.6 | 1.8 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.6 | 6.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.6 | 2.4 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.6 | 4.0 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.6 | 1.7 | GO:0010840 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) circadian sleep/wake cycle, wakefulness(GO:0042746) |
| 0.5 | 2.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.5 | 1.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.5 | 2.7 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.5 | 2.7 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.5 | 3.6 | GO:0030578 | PML body organization(GO:0030578) |
| 0.5 | 1.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.5 | 1.5 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.5 | 2.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.5 | 2.0 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.5 | 12.6 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.5 | 3.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.5 | 1.9 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.5 | 10.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 2.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 3.6 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.4 | 2.2 | GO:0072021 | ascending thin limb development(GO:0072021) thick ascending limb development(GO:0072023) metanephric ascending thin limb development(GO:0072218) metanephric thick ascending limb development(GO:0072233) |
| 0.4 | 1.8 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.4 | 1.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.4 | 5.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 7.3 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.4 | 2.5 | GO:0033625 | positive regulation of integrin activation(GO:0033625) protein secretion by platelet(GO:0070560) |
| 0.4 | 2.9 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 1.7 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.4 | 2.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.6 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.4 | 2.0 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.4 | 0.8 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.4 | 4.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.4 | 1.9 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.4 | 1.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.4 | 3.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.4 | 1.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.3 | 4.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 3.4 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.3 | 4.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 1.7 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 1.7 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.3 | 3.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 7.2 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.3 | 3.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 1.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.3 | 2.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 3.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 3.7 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.3 | 3.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.3 | 1.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.3 | 2.2 | GO:0097106 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.3 | 1.9 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.3 | 0.8 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 | 6.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.5 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.3 | 1.8 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.3 | 2.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 10.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.7 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 0.7 | GO:1900158 | hormone-mediated apoptotic signaling pathway(GO:0008628) positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 0.5 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 1.4 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 9.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 4.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 3.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 0.7 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 1.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.2 | 8.3 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.2 | 1.0 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.2 | 7.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 3.0 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.2 | 3.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.2 | 2.7 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.2 | 5.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.2 | 0.9 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.2 | 2.7 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.2 | 4.7 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 5.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 3.0 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.2 | 5.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 0.9 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.2 | 1.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 3.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.2 | 2.3 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.2 | 0.9 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.2 | 4.8 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 5.9 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 4.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.3 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.6 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.1 | 0.6 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.1 | 0.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 1.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 5.9 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 6.7 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 0.6 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 1.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 2.4 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.1 | 7.2 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 5.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 2.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 1.5 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.1 | 6.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 1.0 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 1.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.5 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 2.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 1.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.7 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 4.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 3.9 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.1 | 2.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.1 | 2.0 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 11.3 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 1.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 3.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 2.5 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.1 | 2.1 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.5 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 9.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 4.0 | GO:0055001 | muscle cell development(GO:0055001) |
| 0.1 | 1.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.1 | 2.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.9 | GO:0010543 | regulation of platelet activation(GO:0010543) |
| 0.1 | 1.9 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.1 | 0.9 | GO:0007595 | lactation(GO:0007595) |
| 0.1 | 3.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 5.2 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 2.7 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) |
| 0.0 | 3.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.8 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 1.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 2.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.0 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 4.0 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 3.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.3 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 1.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 3.8 | GO:1990138 | neuron projection extension(GO:1990138) |
| 0.0 | 0.7 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 2.2 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 1.1 | GO:0048839 | inner ear development(GO:0048839) |
| 0.0 | 1.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 1.4 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 1.6 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 2.3 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.2 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 2.4 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.4 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 2.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 18.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 1.7 | 5.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.5 | 7.4 | GO:0001652 | granular component(GO:0001652) |
| 1.3 | 5.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 1.2 | 6.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 1.1 | 5.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 1.0 | 8.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.9 | 3.6 | GO:0043293 | apoptosome(GO:0043293) |
| 0.9 | 2.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.9 | 12.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.9 | 2.6 | GO:0033011 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.8 | 9.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.8 | 6.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 16.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.7 | 3.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.7 | 6.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 4.3 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.7 | 2.7 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.6 | 2.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.6 | 1.9 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.6 | 16.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.6 | 3.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 3.6 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.5 | 3.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.5 | 2.3 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.4 | 3.4 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 6.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 2.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 9.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.4 | 5.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 5.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 1.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.3 | 26.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 3.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 3.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.2 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 6.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 2.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 20.7 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.2 | 9.3 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 1.7 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 1.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 4.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 3.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 4.7 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 20.3 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 1.8 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.2 | 2.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 3.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 2.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.9 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 11.7 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 4.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 3.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 10.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.6 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 2.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 8.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 5.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 17.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 5.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 20.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 2.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 4.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 2.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.9 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 4.0 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 2.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 5.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 3.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 7.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 5.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.4 | GO:0044215 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 1.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.7 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 2.8 | GO:0045121 | membrane raft(GO:0045121) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 2.5 | 7.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 2.2 | 11.2 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 2.2 | 8.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 2.0 | 7.9 | GO:0030305 | heparanase activity(GO:0030305) |
| 1.9 | 7.7 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 1.9 | 18.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.6 | 4.9 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 1.5 | 4.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 1.2 | 4.7 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 1.1 | 3.4 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 1.1 | 6.7 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 1.1 | 4.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 1.0 | 7.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.0 | 5.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.9 | 12.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.9 | 11.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.9 | 10.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.9 | 2.7 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.9 | 2.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.9 | 5.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.9 | 2.7 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.8 | 5.0 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.8 | 2.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.7 | 3.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.7 | 4.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 2.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.7 | 5.9 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.6 | 3.8 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 1.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.6 | 2.4 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.6 | 5.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.5 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.5 | 1.6 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.5 | 2.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.5 | 2.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.5 | 7.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 5.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.5 | 3.3 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.5 | 3.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 13.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 1.7 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.4 | 2.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 2.1 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.4 | 1.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.4 | 4.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.4 | 7.0 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.4 | 1.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 4.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 2.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 4.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 6.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.3 | 15.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.3 | 6.8 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 1.9 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.3 | 5.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 1.8 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.3 | 3.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 8.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 3.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 4.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 3.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 3.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 5.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.3 | 3.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.3 | 1.5 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 1.3 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.3 | 0.8 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 1.9 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 1.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 2.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 1.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 24.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 0.9 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.2 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 3.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 0.9 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.2 | 4.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 3.6 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.2 | 2.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.2 | 5.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 2.0 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 5.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.2 | 12.6 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 3.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.2 | 3.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 2.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 0.6 | GO:0004040 | amidase activity(GO:0004040) |
| 0.2 | 5.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 0.8 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.2 | 3.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 0.8 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 2.7 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 7.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.7 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 7.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 2.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 4.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 4.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 3.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.8 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.9 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 1.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 5.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 12.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 2.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 3.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 4.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 9.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 9.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.1 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 3.0 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.1 | 0.8 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.1 | 2.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 21.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.7 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 5.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 8.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 8.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 11.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 2.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 1.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 3.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 3.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 6.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 1.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 3.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 3.7 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 1.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 5.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 3.0 | GO:0017016 | Ras GTPase binding(GO:0017016) |
| 0.0 | 4.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.3 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0016248 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 1.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 18.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.6 | 14.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.5 | 5.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 11.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.4 | 6.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 6.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 23.9 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.3 | 22.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.3 | 7.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 10.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.3 | 13.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 40.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 5.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 12.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 6.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 11.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 37.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 4.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 8.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 5.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 5.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.9 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 0.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 8.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 24.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.6 | 10.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.5 | 7.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 5.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.4 | 6.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 8.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 6.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 6.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 3.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.3 | 5.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.3 | 15.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 5.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 4.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 5.8 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.2 | 3.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 7.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 4.0 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.2 | 5.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 4.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 4.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 7.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.8 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 6.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 5.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 6.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 7.3 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 4.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.5 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 2.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 5.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 6.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |