Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
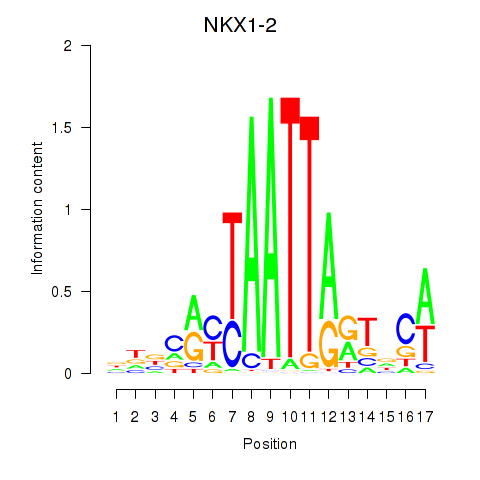
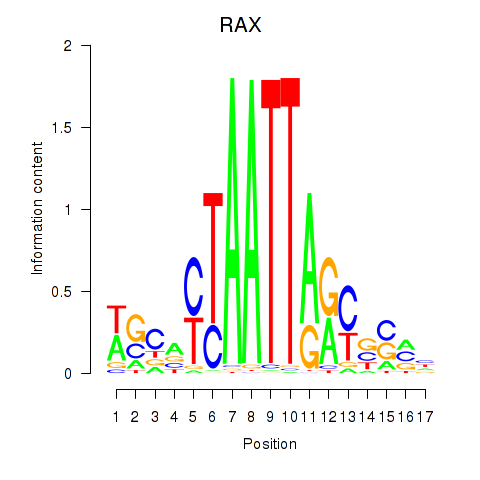
Results for NKX1-2_RAX
Z-value: 0.60
Transcription factors associated with NKX1-2_RAX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-2
|
ENSG00000229544.6 | NK1 homeobox 2 |
|
RAX
|
ENSG00000134438.9 | retina and anterior neural fold homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RAX | hg19_v2_chr18_-_56940611_56940660 | -0.30 | 5.7e-06 | Click! |
Activity profile of NKX1-2_RAX motif
Sorted Z-values of NKX1-2_RAX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_34204642 | 12.29 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_+_187371440 | 9.64 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr7_-_87856280 | 9.57 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr7_-_87856303 | 9.31 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr4_-_25865159 | 9.02 |
ENST00000502949.1
ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr10_+_124914285 | 6.59 |
ENST00000407911.2
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr4_-_103749179 | 5.81 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_103749205 | 5.34 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_23851928 | 4.42 |
ENST00000467766.1
ENST00000424381.1 |
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chrX_-_77225135 | 4.36 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr12_+_32832203 | 4.30 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr10_+_124913930 | 4.30 |
ENST00000368858.5
|
BUB3
|
BUB3 mitotic checkpoint protein |
| chr6_+_30687978 | 4.24 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr3_+_158519654 | 4.17 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr1_+_155278539 | 4.02 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr1_+_155278625 | 3.77 |
ENST00000368356.4
ENST00000356657.6 |
FDPS
|
farnesyl diphosphate synthase |
| chr11_-_111741994 | 3.62 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr2_-_86422095 | 3.52 |
ENST00000254636.5
|
IMMT
|
inner membrane protein, mitochondrial |
| chr3_+_183853052 | 3.24 |
ENST00000273783.3
ENST00000432569.1 ENST00000444495.1 |
EIF2B5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa |
| chr2_-_220264703 | 3.18 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chrX_+_77154935 | 3.14 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr10_+_124913793 | 3.07 |
ENST00000368865.4
ENST00000538238.1 ENST00000368859.2 |
BUB3
|
BUB3 mitotic checkpoint protein |
| chr2_+_90077680 | 2.96 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr4_-_103749313 | 2.75 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_26440700 | 2.33 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr17_-_27418537 | 2.33 |
ENST00000408971.2
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr5_-_82969405 | 2.31 |
ENST00000510978.1
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1 |
| chr19_+_11071546 | 2.29 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr2_-_74619152 | 2.26 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr3_-_105588231 | 2.22 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_+_217524323 | 2.21 |
ENST00000456764.1
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr4_-_103749105 | 2.14 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_+_27008865 | 1.92 |
ENST00000335756.4
ENST00000233505.8 |
CENPA
|
centromere protein A |
| chr4_-_103748880 | 1.84 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_112279782 | 1.83 |
ENST00000550735.2
|
MAPKAPK5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr7_+_100770328 | 1.71 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr3_-_151034734 | 1.62 |
ENST00000260843.4
|
GPR87
|
G protein-coupled receptor 87 |
| chr1_+_43148059 | 1.60 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr7_-_86849883 | 1.50 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr11_+_73498898 | 1.50 |
ENST00000535529.1
ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48
|
mitochondrial ribosomal protein L48 |
| chr11_-_3859089 | 1.41 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr6_+_26402465 | 1.35 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr6_+_26365443 | 1.32 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr2_-_74618964 | 1.24 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr1_+_153746683 | 1.24 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr9_+_125133315 | 1.16 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chrX_+_114827818 | 1.06 |
ENST00000420625.2
|
PLS3
|
plastin 3 |
| chr4_-_66536057 | 1.05 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr8_-_101571933 | 1.03 |
ENST00000520311.1
|
ANKRD46
|
ankyrin repeat domain 46 |
| chr6_-_111927062 | 1.00 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr7_-_99716952 | 1.00 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr8_-_101571964 | 0.88 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr20_-_1447467 | 0.87 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr15_+_64680003 | 0.85 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr19_-_3985455 | 0.82 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chrX_+_19362011 | 0.79 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr5_-_94890648 | 0.79 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr3_-_11685345 | 0.78 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr9_+_125132803 | 0.77 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_+_110736659 | 0.76 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr11_+_5710919 | 0.76 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr7_-_99717463 | 0.75 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_+_138145076 | 0.74 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr20_-_1447547 | 0.72 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr9_+_34646624 | 0.72 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr7_-_87342564 | 0.69 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr4_-_66536196 | 0.69 |
ENST00000511294.1
|
EPHA5
|
EPH receptor A5 |
| chr22_-_32651326 | 0.69 |
ENST00000266086.4
|
SLC5A4
|
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr2_-_89597542 | 0.66 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr8_-_102803163 | 0.66 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr19_+_48949030 | 0.66 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr8_-_42623747 | 0.65 |
ENST00000534622.1
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr11_-_65625678 | 0.61 |
ENST00000308162.5
|
CFL1
|
cofilin 1 (non-muscle) |
| chr22_+_22730353 | 0.61 |
ENST00000390296.2
|
IGLV5-45
|
immunoglobulin lambda variable 5-45 |
| chr10_-_114206649 | 0.61 |
ENST00000369404.3
ENST00000369405.3 |
ZDHHC6
|
zinc finger, DHHC-type containing 6 |
| chr1_+_192605252 | 0.59 |
ENST00000391995.2
ENST00000543215.1 |
RGS13
|
regulator of G-protein signaling 13 |
| chr1_-_67266939 | 0.56 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr1_+_153747746 | 0.52 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr16_-_28608364 | 0.49 |
ENST00000533150.1
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr1_+_186265399 | 0.47 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr12_-_12837423 | 0.47 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr6_+_26402517 | 0.46 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr12_-_10959892 | 0.42 |
ENST00000240615.2
|
TAS2R8
|
taste receptor, type 2, member 8 |
| chr10_-_105845674 | 0.42 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr9_+_34646651 | 0.40 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr7_+_120628731 | 0.36 |
ENST00000310396.5
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_-_82957433 | 0.32 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr3_+_186353756 | 0.28 |
ENST00000431018.1
ENST00000450521.1 ENST00000539949.1 |
FETUB
|
fetuin B |
| chr7_+_50348268 | 0.26 |
ENST00000438033.1
ENST00000439701.1 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr1_+_201979645 | 0.23 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr10_-_92681033 | 0.22 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_+_62439037 | 0.22 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr1_-_151804314 | 0.22 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr12_-_10022735 | 0.20 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr11_-_63376013 | 0.17 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr6_-_111927449 | 0.15 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr9_+_135937365 | 0.13 |
ENST00000372080.4
ENST00000351304.7 |
CEL
|
carboxyl ester lipase |
| chr1_-_190446759 | 0.13 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr16_-_28608424 | 0.05 |
ENST00000335715.4
|
SULT1A2
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 |
| chr8_+_107738240 | 0.05 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr1_+_28261492 | 0.05 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr2_+_74682150 | 0.04 |
ENST00000233331.7
ENST00000431187.1 ENST00000409917.1 ENST00000409493.2 |
INO80B
|
INO80 complex subunit B |
| chr11_-_102651343 | 0.01 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr19_-_46285736 | 0.00 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-2_RAX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 2.7 | 18.9 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 1.6 | 7.8 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 1.4 | 4.3 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 1.1 | 4.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.6 | 1.7 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 4.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 2.3 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.4 | 3.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.4 | 3.2 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.4 | 1.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 16.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.3 | 14.0 | GO:0008608 | mitotic spindle assembly checkpoint(GO:0007094) attachment of spindle microtubules to kinetochore(GO:0008608) spindle assembly checkpoint(GO:0071173) |
| 0.3 | 1.9 | GO:0051382 | kinetochore assembly(GO:0051382) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 1.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 3.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.8 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 3.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 3.5 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 3.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.6 | GO:2000771 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 2.3 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.1 | 0.6 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.8 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.1 | 1.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.7 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.7 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 1.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.8 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 2.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.2 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 4.2 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 0.2 | GO:1904177 | regulation of adipose tissue development(GO:1904177) |
| 0.1 | 1.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.4 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 2.3 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.0 | 0.7 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 3.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.5 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.8 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.0 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 2.7 | 18.9 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 1.1 | 12.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.6 | 1.9 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.5 | 3.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.4 | 3.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.4 | 1.6 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 4.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 3.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.6 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.2 | 0.8 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 3.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 4.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.8 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 2.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.8 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 4.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 11.9 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 3.4 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.2 | 3.6 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.1 | 4.4 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 12.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.7 | 4.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 18.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.5 | 1.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.4 | 17.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 4.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 4.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 2.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 0.8 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 1.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 2.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 3.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 15.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 3.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 3.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 3.2 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.8 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.7 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 9.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 3.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 17.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 11.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 15.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 1.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 12.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 17.4 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.5 | 17.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.3 | 7.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 7.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 4.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 3.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |