Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
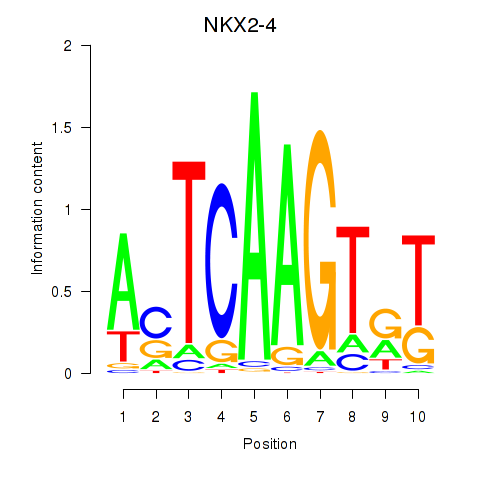
Results for NKX2-4
Z-value: 0.65
Transcription factors associated with NKX2-4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-4
|
ENSG00000125816.4 | NK2 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-4 | hg19_v2_chr20_-_21378666_21378666 | -0.11 | 1.0e-01 | Click! |
Activity profile of NKX2-4 motif
Sorted Z-values of NKX2-4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_49066811 | 10.35 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr10_-_28571015 | 7.60 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr2_-_225434538 | 7.02 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr1_-_153643442 | 6.46 |
ENST00000368681.1
ENST00000361891.4 |
ILF2
|
interleukin enhancer binding factor 2 |
| chr7_-_96339132 | 6.39 |
ENST00000413065.1
|
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr7_-_93519471 | 6.34 |
ENST00000451238.1
|
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr7_+_116139424 | 6.18 |
ENST00000222693.4
|
CAV2
|
caveolin 2 |
| chr11_+_62439126 | 6.07 |
ENST00000377953.3
|
C11orf83
|
chromosome 11 open reading frame 83 |
| chr7_+_75932863 | 6.04 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr17_-_38574169 | 5.91 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr12_+_8995832 | 5.75 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr15_+_63334831 | 5.42 |
ENST00000288398.6
ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1
|
tropomyosin 1 (alpha) |
| chr17_-_15587602 | 5.38 |
ENST00000416464.2
ENST00000578237.1 ENST00000581200.1 |
TRIM16
|
tripartite motif containing 16 |
| chr15_+_80364901 | 5.34 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr18_-_33702078 | 5.09 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_-_32022733 | 5.03 |
ENST00000438237.2
ENST00000396556.2 |
OSBPL10
|
oxysterol binding protein-like 10 |
| chr6_+_83072923 | 4.98 |
ENST00000535040.1
|
TPBG
|
trophoblast glycoprotein |
| chr6_+_83073334 | 4.89 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr8_-_74206673 | 4.79 |
ENST00000396465.1
|
RPL7
|
ribosomal protein L7 |
| chr15_-_63448973 | 4.68 |
ENST00000462430.1
|
RPS27L
|
ribosomal protein S27-like |
| chr11_+_118958689 | 4.21 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr17_-_5138099 | 4.19 |
ENST00000571800.1
ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr4_-_103746683 | 4.16 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_81272287 | 3.98 |
ENST00000520547.2
|
EIF5AL1
|
eukaryotic translation initiation factor 5A-like 1 |
| chr19_+_37998031 | 3.91 |
ENST00000586138.1
ENST00000588578.1 ENST00000587986.1 |
ZNF793
|
zinc finger protein 793 |
| chr12_+_96252706 | 3.85 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr13_+_53216565 | 3.84 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr7_-_96339167 | 3.71 |
ENST00000444799.1
ENST00000417009.1 ENST00000248566.2 |
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr15_-_34394008 | 3.64 |
ENST00000527822.1
ENST00000528949.1 |
EMC7
|
ER membrane protein complex subunit 7 |
| chr8_+_104310661 | 3.62 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr7_+_134551583 | 3.61 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr14_+_56078695 | 3.55 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr11_+_60223312 | 3.54 |
ENST00000532491.1
ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr7_-_54826869 | 3.52 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr11_+_60223225 | 3.40 |
ENST00000524807.1
ENST00000345732.4 |
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr14_+_62229075 | 3.35 |
ENST00000216294.4
|
SNAPC1
|
small nuclear RNA activating complex, polypeptide 1, 43kDa |
| chr4_-_103747011 | 3.33 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_-_156721502 | 3.21 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr14_-_23451467 | 3.20 |
ENST00000555074.1
ENST00000361265.4 |
RP11-298I3.5
AJUBA
|
RP11-298I3.5 ajuba LIM protein |
| chr1_-_94147385 | 3.16 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr11_+_844406 | 3.16 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr10_+_105005644 | 3.08 |
ENST00000441178.2
|
RP11-332O19.5
|
ribulose-5-phosphate-3-epimerase-like 1 |
| chr4_-_103746924 | 3.07 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_+_169418195 | 3.06 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr19_-_41942344 | 3.05 |
ENST00000594660.1
|
ATP5SL
|
ATP5S-like |
| chr8_-_49833978 | 3.04 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr15_-_34394119 | 3.01 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr12_-_57113333 | 2.99 |
ENST00000550920.1
|
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chr2_-_188419200 | 2.95 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr11_-_85780853 | 2.91 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr12_+_54422142 | 2.87 |
ENST00000243108.4
|
HOXC6
|
homeobox C6 |
| chr5_-_98262240 | 2.81 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr5_-_34919094 | 2.79 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr11_-_14521379 | 2.71 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr2_-_179315786 | 2.67 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr7_-_142232071 | 2.64 |
ENST00000390364.3
|
TRBV10-1
|
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr14_-_23058063 | 2.58 |
ENST00000538631.1
ENST00000543337.1 ENST00000250498.4 |
DAD1
|
defender against cell death 1 |
| chr7_+_129932974 | 2.56 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr8_+_97597148 | 2.56 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr12_+_124069070 | 2.54 |
ENST00000262225.3
ENST00000438031.2 |
TMED2
|
transmembrane emp24 domain trafficking protein 2 |
| chr12_+_10658201 | 2.47 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr1_-_217250231 | 2.47 |
ENST00000493748.1
ENST00000463665.1 |
ESRRG
|
estrogen-related receptor gamma |
| chr6_-_131277510 | 2.45 |
ENST00000525193.1
ENST00000527659.1 |
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr3_-_107777208 | 2.41 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr10_+_22605374 | 2.38 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr4_+_39699664 | 2.37 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chrX_+_49294472 | 2.34 |
ENST00000361446.5
|
GAGE12B
|
G antigen 12B |
| chr2_+_28615669 | 2.32 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr9_+_131445928 | 2.26 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr11_-_62439012 | 2.25 |
ENST00000532208.1
ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48
|
chromosome 11 open reading frame 48 |
| chr14_+_64680854 | 2.25 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr9_+_90341024 | 2.24 |
ENST00000340342.6
ENST00000342020.5 |
CTSL
|
cathepsin L |
| chr1_-_95391315 | 2.23 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr2_-_37899323 | 2.22 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr14_+_24702099 | 2.20 |
ENST00000420554.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chr11_+_65769946 | 2.14 |
ENST00000533166.1
|
BANF1
|
barrier to autointegration factor 1 |
| chr14_-_71067360 | 2.13 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chr7_+_29234375 | 2.12 |
ENST00000409350.1
ENST00000495789.2 ENST00000539389.1 |
CHN2
|
chimerin 2 |
| chr8_-_141774467 | 2.12 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr4_+_75480629 | 2.10 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr6_-_149969871 | 2.09 |
ENST00000335643.8
ENST00000444282.1 |
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr2_-_219925189 | 2.08 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr2_+_161993412 | 2.07 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_22605304 | 2.05 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr9_-_127177703 | 2.05 |
ENST00000259457.3
ENST00000536392.1 ENST00000441097.1 |
PSMB7
|
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr12_-_56843161 | 2.02 |
ENST00000554616.1
ENST00000553532.1 ENST00000229201.4 |
TIMELESS
|
timeless circadian clock |
| chr5_+_159436120 | 2.00 |
ENST00000522793.1
ENST00000231238.5 |
TTC1
|
tetratricopeptide repeat domain 1 |
| chr17_-_26684473 | 1.99 |
ENST00000540200.1
|
POLDIP2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr6_-_149969829 | 1.98 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr2_-_27435125 | 1.98 |
ENST00000414408.1
ENST00000310574.3 |
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr11_+_65769550 | 1.92 |
ENST00000312175.2
ENST00000445560.2 ENST00000530204.1 |
BANF1
|
barrier to autointegration factor 1 |
| chrX_+_100075368 | 1.92 |
ENST00000415585.2
ENST00000372972.2 ENST00000413437.1 |
CSTF2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
| chr2_-_188419078 | 1.91 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr12_+_123011776 | 1.90 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr11_-_19223523 | 1.87 |
ENST00000265968.3
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr2_-_176046391 | 1.85 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr17_+_66244071 | 1.82 |
ENST00000580548.1
ENST00000580753.1 ENST00000392720.2 ENST00000359783.4 ENST00000584837.1 ENST00000579724.1 ENST00000584494.1 ENST00000580837.1 |
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr12_-_57119106 | 1.79 |
ENST00000546392.1
|
NACA
|
nascent polypeptide-associated complex alpha subunit |
| chrX_-_119445306 | 1.74 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr10_+_35484053 | 1.72 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chrX_+_49178536 | 1.69 |
ENST00000442437.2
|
GAGE12J
|
G antigen 12J |
| chr10_-_11574274 | 1.68 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr7_+_120629653 | 1.66 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr11_-_85779971 | 1.66 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr2_-_37544209 | 1.66 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr11_+_34642656 | 1.66 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr14_+_74353320 | 1.65 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr14_+_24701819 | 1.65 |
ENST00000560139.1
ENST00000559910.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr8_-_141810634 | 1.63 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chrX_+_78200829 | 1.60 |
ENST00000544091.1
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr1_-_70671216 | 1.56 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr6_+_31674639 | 1.55 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr1_+_32042105 | 1.55 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chr1_+_148560843 | 1.54 |
ENST00000442702.2
ENST00000369187.3 |
NBPF15
|
neuroblastoma breakpoint family, member 15 |
| chr1_-_89458287 | 1.53 |
ENST00000370485.2
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr1_+_15986364 | 1.51 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr14_+_24702127 | 1.51 |
ENST00000557854.1
ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr2_+_27435179 | 1.50 |
ENST00000606999.1
ENST00000405489.3 |
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr14_+_24702073 | 1.49 |
ENST00000399440.2
|
GMPR2
|
guanosine monophosphate reductase 2 |
| chrX_-_133792480 | 1.47 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr19_-_20748541 | 1.47 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr7_+_65552756 | 1.47 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr10_+_21823079 | 1.46 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr2_+_37571845 | 1.45 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr9_-_86593238 | 1.44 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chrX_+_78200913 | 1.42 |
ENST00000171757.2
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10 |
| chr5_-_176836577 | 1.39 |
ENST00000253496.3
|
F12
|
coagulation factor XII (Hageman factor) |
| chr10_-_121296045 | 1.38 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr15_+_67430339 | 1.38 |
ENST00000439724.3
|
SMAD3
|
SMAD family member 3 |
| chr8_+_7705398 | 1.33 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr8_-_7320974 | 1.29 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr12_+_58176525 | 1.28 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr2_-_220025548 | 1.28 |
ENST00000356853.5
|
NHEJ1
|
nonhomologous end-joining factor 1 |
| chr12_+_123237321 | 1.28 |
ENST00000280557.6
ENST00000455982.2 |
DENR
|
density-regulated protein |
| chr4_+_109541772 | 1.26 |
ENST00000506397.1
ENST00000394668.2 |
RPL34
|
ribosomal protein L34 |
| chr2_-_166651191 | 1.26 |
ENST00000392701.3
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr11_+_64018955 | 1.25 |
ENST00000279230.6
ENST00000540288.1 ENST00000325234.5 |
PLCB3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr14_+_24701628 | 1.24 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chr14_-_24701539 | 1.24 |
ENST00000534348.1
ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1
NEDD8
|
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr5_+_175298573 | 1.22 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr18_+_55816546 | 1.21 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_+_36235964 | 1.20 |
ENST00000587708.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr9_-_95298314 | 1.18 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr2_+_161993465 | 1.16 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr18_-_28622699 | 1.15 |
ENST00000360428.4
|
DSC3
|
desmocollin 3 |
| chr16_+_28943260 | 1.15 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr11_+_57105991 | 1.14 |
ENST00000263314.2
|
P2RX3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr5_+_150040403 | 1.14 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr19_+_58898627 | 1.14 |
ENST00000598098.1
ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5
|
ribosomal protein S5 |
| chr2_-_170219079 | 1.14 |
ENST00000263816.3
|
LRP2
|
low density lipoprotein receptor-related protein 2 |
| chr7_-_86849025 | 1.13 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr2_+_1417228 | 1.11 |
ENST00000382269.3
ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO
|
thyroid peroxidase |
| chr19_-_55669093 | 1.10 |
ENST00000344887.5
|
TNNI3
|
troponin I type 3 (cardiac) |
| chr6_+_153552455 | 1.10 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr12_+_50451331 | 1.09 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr19_-_49258606 | 1.09 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr17_-_63822563 | 1.08 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr9_-_95166884 | 1.08 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr18_+_56338618 | 1.07 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr3_-_88108192 | 1.07 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr2_-_175462456 | 1.05 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr18_-_28622774 | 1.04 |
ENST00000434452.1
|
DSC3
|
desmocollin 3 |
| chr10_+_21823243 | 1.03 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_197036364 | 1.02 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr9_-_95166841 | 1.02 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr11_-_85780086 | 1.02 |
ENST00000532317.1
ENST00000528256.1 ENST00000526033.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr4_-_155533787 | 1.02 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr11_+_76813886 | 1.01 |
ENST00000529803.1
|
OMP
|
olfactory marker protein |
| chr5_+_175298674 | 1.01 |
ENST00000514150.1
|
CPLX2
|
complexin 2 |
| chr11_+_19138670 | 1.00 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr11_-_19263145 | 0.99 |
ENST00000532666.1
ENST00000527884.1 |
E2F8
|
E2F transcription factor 8 |
| chr1_-_110155671 | 0.97 |
ENST00000351050.3
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr7_-_86849883 | 0.96 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr3_-_126327398 | 0.96 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr6_-_31088214 | 0.94 |
ENST00000376288.2
|
CDSN
|
corneodesmosin |
| chr5_+_137203557 | 0.93 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr14_+_22962898 | 0.92 |
ENST00000390492.1
|
TRAJ45
|
T cell receptor alpha joining 45 |
| chrX_-_124097620 | 0.91 |
ENST00000371130.3
ENST00000422452.2 |
TENM1
|
teneurin transmembrane protein 1 |
| chr11_+_12766583 | 0.90 |
ENST00000361985.2
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr1_+_144989309 | 0.90 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr3_-_88108212 | 0.89 |
ENST00000482016.1
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr14_+_22356029 | 0.88 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2 |
| chr16_+_81070792 | 0.88 |
ENST00000564241.1
ENST00000565237.1 |
ATMIN
|
ATM interactor |
| chr6_+_5261225 | 0.87 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr17_+_75277492 | 0.86 |
ENST00000427177.1
ENST00000591198.1 |
SEPT9
|
septin 9 |
| chr16_+_3062457 | 0.86 |
ENST00000445369.2
|
CLDN9
|
claudin 9 |
| chr8_+_22429205 | 0.85 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr10_-_69455873 | 0.85 |
ENST00000433211.2
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3 |
| chr1_+_152627927 | 0.84 |
ENST00000444515.1
ENST00000536536.1 |
LINC00302
|
long intergenic non-protein coding RNA 302 |
| chr3_+_118905564 | 0.84 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chrX_-_74376108 | 0.83 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr1_+_161136180 | 0.83 |
ENST00000352210.5
ENST00000367999.4 ENST00000544598.1 ENST00000535223.1 ENST00000432542.2 |
PPOX
|
protoporphyrinogen oxidase |
| chr1_+_13359819 | 0.83 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr18_+_66465302 | 0.83 |
ENST00000360242.5
ENST00000358653.5 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr14_-_104028595 | 0.82 |
ENST00000337322.4
ENST00000445922.2 |
BAG5
|
BCL2-associated athanogene 5 |
| chr12_-_123011536 | 0.81 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chrX_-_110655306 | 0.81 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr6_-_136610911 | 0.80 |
ENST00000530767.1
ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1
|
BCL2-associated transcription factor 1 |
| chr12_-_123011476 | 0.79 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr13_+_49280951 | 0.77 |
ENST00000282018.3
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 1.7 | 10.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.5 | 6.0 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 1.1 | 7.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 1.0 | 6.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 1.0 | 3.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.9 | 3.6 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.8 | 3.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.8 | 5.6 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.7 | 2.8 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.7 | 1.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.7 | 4.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.7 | 5.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.6 | 2.5 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.6 | 4.9 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.6 | 5.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.6 | 5.9 | GO:0030263 | resolution of meiotic recombination intermediates(GO:0000712) apoptotic chromosome condensation(GO:0030263) embryonic cleavage(GO:0040016) |
| 0.6 | 3.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.5 | 3.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.5 | 2.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.5 | 5.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.5 | 4.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.5 | 1.4 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.5 | 1.9 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.4 | 2.7 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.4 | 1.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 3.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 4.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.4 | 2.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.4 | 5.0 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.4 | 2.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.4 | 2.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.4 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.4 | 1.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.3 | 4.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 1.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 1.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 2.4 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.3 | 1.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 6.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.3 | 6.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) purine-containing compound salvage(GO:0043101) |
| 0.3 | 0.8 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.2 | 1.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 5.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.7 | GO:0002661 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.2 | 5.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.2 | 0.9 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.6 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 2.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 0.6 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 2.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 11.0 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.2 | 1.0 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.2 | 2.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.2 | 0.5 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 3.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 6.2 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.2 | 2.5 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.2 | 2.2 | GO:0071888 | macrophage apoptotic process(GO:0071888) |
| 0.1 | 0.4 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.1 | 0.8 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 | 2.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 2.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.4 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.6 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 1.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.8 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 2.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 3.3 | GO:0072663 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.9 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.4 | GO:1902941 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) |
| 0.1 | 1.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 8.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 3.8 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.3 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 2.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.8 | GO:0002005 | angiotensin catabolic process in blood(GO:0002005) |
| 0.1 | 0.3 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.1 | 1.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.8 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 1.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 0.5 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 1.5 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.1 | 1.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.6 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.6 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.1 | 2.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 1.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 1.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 1.5 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 1.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 4.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.6 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.5 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.5 | GO:0048840 | otolith development(GO:0048840) |
| 0.1 | 6.7 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.2 | GO:0031293 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 5.9 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.1 | 0.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 1.0 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 1.1 | GO:0001976 | neurological system process involved in regulation of systemic arterial blood pressure(GO:0001976) |
| 0.1 | 0.4 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 1.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 2.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 2.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.3 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.8 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 1.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 2.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 2.2 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 3.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.9 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.9 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 3.0 | GO:0007127 | meiosis I(GO:0007127) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.8 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 1.5 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.4 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 2.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.6 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0045636 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) |
| 0.0 | 2.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 1.4 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 2.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 2.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 2.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 1.3 | 8.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.9 | 5.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.8 | 7.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.7 | 6.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.7 | 2.8 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.7 | 4.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.6 | 7.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.5 | 3.8 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.5 | 1.9 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.4 | 4.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 2.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.3 | 2.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.3 | 5.4 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 3.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 2.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 6.2 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 1.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 7.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 1.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 0.9 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 2.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 1.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 12.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.9 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 2.0 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.1 | 6.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 1.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 3.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 3.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 4.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 6.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 5.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.4 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 12.3 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.3 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 6.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 3.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 4.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.8 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 7.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 10.4 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 3.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 4.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 2.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 9.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 3.8 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 3.0 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.0 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 8.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.8 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.9 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 2.6 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 10.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 2.0 | 6.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 2.0 | 5.9 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 1.8 | 5.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 1.8 | 7.0 | GO:0031208 | POZ domain binding(GO:0031208) |
| 1.3 | 8.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.7 | 4.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.7 | 2.0 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.6 | 3.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.5 | 1.4 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.5 | 5.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 2.8 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.4 | 1.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 1.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 2.5 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.4 | 3.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 3.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.3 | 2.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.3 | 1.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.3 | 4.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 5.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.6 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 3.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 2.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 2.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 11.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 6.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 3.7 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 0.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.6 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 3.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 5.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 4.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 2.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 1.0 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.2 | 0.7 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.2 | 3.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 2.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.8 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 3.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.9 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 15.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 2.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 2.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 2.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.4 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 11.7 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 5.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 5.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.4 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 2.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 2.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 6.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.7 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 1.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.8 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 1.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 3.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 0.4 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 4.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 8.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 2.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 6.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 3.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 3.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 11.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 7.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 8.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 8.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 6.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 7.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 5.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 7.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 9.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 2.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 3.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 3.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 10.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.5 | 4.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 5.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 6.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.3 | 2.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.3 | 10.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 3.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 7.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.2 | 3.7 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 5.0 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.5 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.2 | 3.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 9.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.9 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 8.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 1.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.9 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 5.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 7.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.0 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 3.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 1.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |