Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
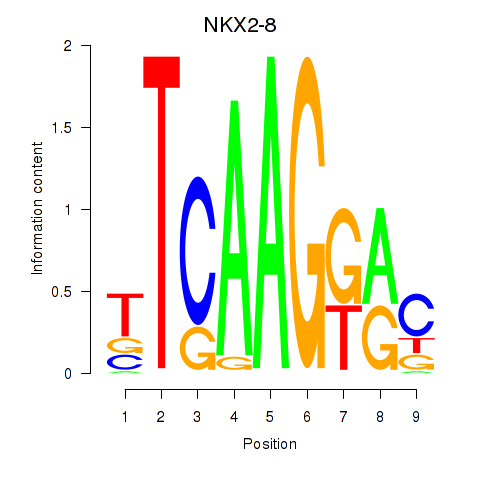
Results for NKX2-8
Z-value: 0.23
Transcription factors associated with NKX2-8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-8
|
ENSG00000136327.6 | NK2 homeobox 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-8 | hg19_v2_chr14_-_37051798_37051831 | -0.28 | 3.3e-05 | Click! |
Activity profile of NKX2-8 motif
Sorted Z-values of NKX2-8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_107049312 | 8.59 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr14_-_106573756 | 7.98 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr14_-_107131560 | 7.71 |
ENST00000390632.2
|
IGHV3-66
|
immunoglobulin heavy variable 3-66 |
| chr14_-_106994333 | 6.88 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr14_-_106963409 | 6.82 |
ENST00000390621.2
|
IGHV1-45
|
immunoglobulin heavy variable 1-45 |
| chr3_-_58613323 | 6.38 |
ENST00000474531.1
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107, member A |
| chr14_-_106552755 | 6.25 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr14_-_106586656 | 6.07 |
ENST00000390602.2
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr14_-_106692191 | 5.81 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr14_-_107219365 | 5.79 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr15_-_20170354 | 5.30 |
ENST00000338912.5
|
IGHV1OR15-9
|
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr14_-_106845789 | 5.02 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr14_-_106518922 | 4.83 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr2_-_55277436 | 4.76 |
ENST00000354474.6
|
RTN4
|
reticulon 4 |
| chr16_-_33647696 | 4.73 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr2_-_55277512 | 4.65 |
ENST00000402434.2
|
RTN4
|
reticulon 4 |
| chr8_-_81083341 | 4.51 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr12_+_25205568 | 4.47 |
ENST00000548766.1
ENST00000556887.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr15_-_77363441 | 4.40 |
ENST00000346495.2
ENST00000424443.3 ENST00000561277.1 |
TSPAN3
|
tetraspanin 3 |
| chr2_-_55277692 | 4.39 |
ENST00000394611.2
|
RTN4
|
reticulon 4 |
| chr2_-_55277654 | 4.35 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr5_-_78808617 | 4.29 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr15_-_77363375 | 4.21 |
ENST00000559494.1
|
TSPAN3
|
tetraspanin 3 |
| chr15_-_77363513 | 4.14 |
ENST00000267970.4
|
TSPAN3
|
tetraspanin 3 |
| chr12_-_21810726 | 3.86 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr12_-_21810765 | 3.85 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr16_+_32063311 | 3.83 |
ENST00000426099.1
|
AC142381.1
|
AC142381.1 |
| chr16_+_32077386 | 3.83 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr5_-_176056974 | 3.81 |
ENST00000510387.1
ENST00000506696.1 |
SNCB
|
synuclein, beta |
| chr3_+_111717600 | 3.47 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr2_+_136289030 | 3.35 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr3_-_16524357 | 3.33 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr12_+_25205666 | 3.31 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr16_+_33020496 | 3.22 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr6_-_32731299 | 3.09 |
ENST00000435145.2
ENST00000437316.2 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr3_+_111717511 | 3.03 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr2_-_132249955 | 2.96 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr14_-_106791536 | 2.80 |
ENST00000390613.2
|
IGHV3-30
|
immunoglobulin heavy variable 3-30 |
| chr16_+_88872176 | 2.61 |
ENST00000569140.1
|
CDT1
|
chromatin licensing and DNA replication factor 1 |
| chr16_+_33006369 | 2.52 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr16_+_33629600 | 2.23 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr12_+_25205446 | 2.22 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr15_+_51973680 | 2.14 |
ENST00000542355.2
|
SCG3
|
secretogranin III |
| chr6_+_32146131 | 2.09 |
ENST00000375094.3
|
RNF5
|
ring finger protein 5, E3 ubiquitin protein ligase |
| chr12_+_13349650 | 2.09 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr10_+_118305435 | 2.04 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase |
| chr12_+_51442101 | 1.99 |
ENST00000550929.1
ENST00000262055.4 ENST00000550442.1 ENST00000549340.1 ENST00000548209.1 ENST00000548251.1 ENST00000550814.1 ENST00000547660.1 ENST00000380123.2 ENST00000548401.1 ENST00000418425.2 ENST00000547008.1 ENST00000552739.1 |
LETMD1
|
LETM1 domain containing 1 |
| chr3_-_53290016 | 1.99 |
ENST00000423525.2
ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT
|
transketolase |
| chr15_+_51973550 | 1.98 |
ENST00000220478.3
|
SCG3
|
secretogranin III |
| chr2_+_101591314 | 1.98 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr11_-_77348796 | 1.98 |
ENST00000263309.3
ENST00000525064.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chrX_+_77154935 | 1.94 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr14_-_106622419 | 1.93 |
ENST00000390604.2
|
IGHV3-16
|
immunoglobulin heavy variable 3-16 (non-functional) |
| chr7_+_99816859 | 1.92 |
ENST00000317271.2
|
PVRIG
|
poliovirus receptor related immunoglobulin domain containing |
| chr7_+_30634297 | 1.90 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr2_+_79252804 | 1.87 |
ENST00000393897.2
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr2_+_79252822 | 1.87 |
ENST00000272324.5
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr6_-_32731243 | 1.84 |
ENST00000427449.1
ENST00000411527.1 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr19_+_34856141 | 1.83 |
ENST00000586425.1
|
GPI
|
glucose-6-phosphate isomerase |
| chr2_+_79252834 | 1.79 |
ENST00000409471.1
|
REG3G
|
regenerating islet-derived 3 gamma |
| chr11_-_77348821 | 1.63 |
ENST00000528364.1
ENST00000532069.1 ENST00000525428.1 |
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A |
| chr7_-_766879 | 1.61 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr15_-_75248954 | 1.59 |
ENST00000499788.2
|
RPP25
|
ribonuclease P/MRP 25kDa subunit |
| chr2_-_207024233 | 1.58 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_179050512 | 1.57 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr1_+_158223923 | 1.57 |
ENST00000289429.5
|
CD1A
|
CD1a molecule |
| chr6_+_69942298 | 1.53 |
ENST00000238918.8
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr16_+_22501658 | 1.47 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr11_-_72353451 | 1.46 |
ENST00000376450.3
|
PDE2A
|
phosphodiesterase 2A, cGMP-stimulated |
| chr13_-_46964177 | 1.35 |
ENST00000389908.3
|
KIAA0226L
|
KIAA0226-like |
| chr14_-_58894223 | 1.33 |
ENST00000555593.1
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr13_-_23949671 | 1.32 |
ENST00000402364.1
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr17_+_76210267 | 1.28 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr15_+_93749295 | 1.25 |
ENST00000599897.1
|
AC112693.2
|
AC112693.2 |
| chr1_-_95007193 | 1.23 |
ENST00000370207.4
ENST00000334047.7 |
F3
|
coagulation factor III (thromboplastin, tissue factor) |
| chr1_-_27998689 | 1.21 |
ENST00000339145.4
ENST00000362020.4 ENST00000361157.6 |
IFI6
|
interferon, alpha-inducible protein 6 |
| chr22_-_50523760 | 1.14 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr15_+_40453204 | 1.14 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr3_-_47950745 | 1.11 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr17_-_46178527 | 1.09 |
ENST00000393408.3
|
CBX1
|
chromobox homolog 1 |
| chr16_-_30441293 | 1.06 |
ENST00000565758.1
ENST00000567983.1 ENST00000319285.4 |
DCTPP1
|
dCTP pyrophosphatase 1 |
| chr14_-_24732403 | 1.06 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr5_+_92919043 | 1.01 |
ENST00000327111.3
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr17_-_62502399 | 1.00 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr17_-_62502639 | 1.00 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_+_152957707 | 0.99 |
ENST00000368762.1
|
SPRR1A
|
small proline-rich protein 1A |
| chr7_-_137686791 | 0.98 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr3_-_114343039 | 0.97 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_207024306 | 0.97 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_-_67266939 | 0.95 |
ENST00000304526.2
|
INSL5
|
insulin-like 5 |
| chr11_-_10828892 | 0.95 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_55464600 | 0.91 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr14_+_24540046 | 0.90 |
ENST00000397016.2
ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6
|
copine VI (neuronal) |
| chr3_-_114790179 | 0.89 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr16_-_18468926 | 0.82 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr8_-_141810634 | 0.81 |
ENST00000521986.1
ENST00000523539.1 ENST00000538769.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr15_-_35047166 | 0.78 |
ENST00000290374.4
|
GJD2
|
gap junction protein, delta 2, 36kDa |
| chr1_-_68698222 | 0.75 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr3_+_4535025 | 0.74 |
ENST00000302640.8
ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr5_-_95158644 | 0.72 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chrX_-_64196376 | 0.71 |
ENST00000447788.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr15_+_45422178 | 0.70 |
ENST00000389037.3
ENST00000558322.1 |
DUOX1
|
dual oxidase 1 |
| chr11_-_114466477 | 0.70 |
ENST00000375478.3
|
NXPE4
|
neurexophilin and PC-esterase domain family, member 4 |
| chr17_-_202579 | 0.67 |
ENST00000577079.1
ENST00000331302.7 ENST00000536489.2 |
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chrX_-_64196351 | 0.65 |
ENST00000374839.3
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chrX_-_64196307 | 0.65 |
ENST00000545618.1
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr12_-_11548496 | 0.65 |
ENST00000389362.4
ENST00000565533.1 ENST00000546254.1 |
PRB2
PRB1
|
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr3_-_116163830 | 0.64 |
ENST00000333617.4
|
LSAMP
|
limbic system-associated membrane protein |
| chrX_-_110039286 | 0.61 |
ENST00000434224.1
|
CHRDL1
|
chordin-like 1 |
| chr6_-_132272504 | 0.61 |
ENST00000367976.3
|
CTGF
|
connective tissue growth factor |
| chr16_+_58426296 | 0.61 |
ENST00000426538.2
ENST00000328514.7 ENST00000318129.5 |
GINS3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr6_-_31830655 | 0.61 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr8_+_27182862 | 0.60 |
ENST00000521164.1
ENST00000346049.5 |
PTK2B
|
protein tyrosine kinase 2 beta |
| chr20_-_44485835 | 0.59 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr14_-_24732368 | 0.56 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr10_+_71078595 | 0.55 |
ENST00000359426.6
|
HK1
|
hexokinase 1 |
| chr17_+_76210367 | 0.54 |
ENST00000592734.1
ENST00000587746.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr8_+_104310661 | 0.54 |
ENST00000522566.1
|
FZD6
|
frizzled family receptor 6 |
| chr1_+_42922173 | 0.50 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr7_+_135242652 | 0.49 |
ENST00000285968.6
ENST00000440390.2 |
NUP205
|
nucleoporin 205kDa |
| chr12_+_121837905 | 0.47 |
ENST00000392465.3
ENST00000554606.1 ENST00000392464.2 ENST00000555076.1 |
RNF34
|
ring finger protein 34, E3 ubiquitin protein ligase |
| chr1_-_115292591 | 0.46 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr8_+_67341239 | 0.44 |
ENST00000320270.2
|
RRS1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr21_-_35899113 | 0.42 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr2_-_207023918 | 0.39 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_+_36486078 | 0.37 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chrX_-_48858667 | 0.35 |
ENST00000376423.4
ENST00000376441.1 |
GRIPAP1
|
GRIP1 associated protein 1 |
| chr1_+_32671236 | 0.35 |
ENST00000537469.1
ENST00000291358.6 |
IQCC
|
IQ motif containing C |
| chr19_-_38720354 | 0.35 |
ENST00000416611.1
|
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr3_-_8811288 | 0.34 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr14_-_58894332 | 0.33 |
ENST00000395159.2
|
TIMM9
|
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr17_+_73472575 | 0.30 |
ENST00000375248.5
|
KIAA0195
|
KIAA0195 |
| chr1_-_104238912 | 0.29 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr2_-_207024134 | 0.29 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_+_224301787 | 0.27 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chrX_+_48681768 | 0.27 |
ENST00000430858.1
|
HDAC6
|
histone deacetylase 6 |
| chr8_+_24241789 | 0.26 |
ENST00000256412.4
ENST00000538205.1 |
ADAMDEC1
|
ADAM-like, decysin 1 |
| chr16_-_3767551 | 0.25 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr3_+_173302222 | 0.25 |
ENST00000361589.4
|
NLGN1
|
neuroligin 1 |
| chr13_-_46756351 | 0.25 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr16_+_8807419 | 0.25 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr20_-_48530230 | 0.24 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr1_-_94147385 | 0.24 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_12040238 | 0.23 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr5_+_43602750 | 0.22 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_+_23430157 | 0.22 |
ENST00000399967.3
|
FP15737
|
FP15737 |
| chrX_+_123094369 | 0.21 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr11_-_107590383 | 0.21 |
ENST00000525934.1
ENST00000531293.1 |
SLN
|
sarcolipin |
| chr16_-_3767506 | 0.21 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr14_-_107199464 | 0.20 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr16_-_66952779 | 0.16 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr19_-_18391708 | 0.15 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr6_+_125524785 | 0.15 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr12_+_66218598 | 0.15 |
ENST00000541363.1
|
HMGA2
|
high mobility group AT-hook 2 |
| chr6_+_143772060 | 0.15 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr22_+_38609538 | 0.14 |
ENST00000407965.1
|
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr6_-_32145861 | 0.13 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr1_+_151512775 | 0.13 |
ENST00000368849.3
ENST00000392712.3 ENST00000353024.3 ENST00000368848.2 ENST00000538902.1 |
TUFT1
|
tuftelin 1 |
| chr4_-_110723134 | 0.13 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr10_+_115674530 | 0.13 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr4_-_153601136 | 0.13 |
ENST00000504064.1
ENST00000304385.3 |
TMEM154
|
transmembrane protein 154 |
| chr12_+_121416489 | 0.12 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr16_-_66952742 | 0.12 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr10_-_15762124 | 0.12 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr12_-_371994 | 0.11 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr10_-_118032979 | 0.11 |
ENST00000355422.6
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr1_+_16348525 | 0.10 |
ENST00000331433.4
|
CLCNKA
|
chloride channel, voltage-sensitive Ka |
| chr14_-_36990354 | 0.10 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr3_-_114343768 | 0.08 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr10_-_118032697 | 0.06 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr2_+_119699742 | 0.05 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr4_+_144303093 | 0.05 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr20_+_47835884 | 0.05 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr7_+_112063192 | 0.04 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr7_-_86974767 | 0.03 |
ENST00000610086.1
|
TP53TG1
|
TP53 target 1 (non-protein coding) |
| chrX_-_110654147 | 0.03 |
ENST00000358070.4
|
DCX
|
doublecortin |
| chr2_-_152118352 | 0.03 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr19_-_44259053 | 0.02 |
ENST00000601170.1
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chrX_-_53461288 | 0.02 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr19_-_44259136 | 0.02 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr5_+_149737202 | 0.01 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 18.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.8 | 5.5 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 1.4 | 4.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.9 | 2.6 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.7 | 49.6 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.7 | 2.0 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 1.8 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.5 | 1.6 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.5 | 7.7 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.5 | 1.5 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.5 | 3.3 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.5 | 1.9 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.4 | 43.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 1.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 | 1.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 0.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 1.1 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.2 | 0.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.2 | 1.0 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.2 | 0.7 | GO:0042335 | cuticle development(GO:0042335) |
| 0.2 | 0.5 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.2 | 0.6 | GO:0048749 | compound eye development(GO:0048749) |
| 0.2 | 0.6 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 2.0 | GO:0098856 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.1 | 0.5 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 2.0 | GO:1903800 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 2.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.6 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 3.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.5 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 2.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 2.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.8 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 1.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.1 | 1.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 1.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.5 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 9.6 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 1.1 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.1 | 0.2 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.4 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 4.1 | GO:0042417 | dopamine metabolic process(GO:0042417) |
| 0.1 | 1.9 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.6 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 1.8 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.7 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 1.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 4.9 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 1.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.9 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.6 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.0 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.0 | 5.5 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 49.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.9 | 18.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.5 | 2.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.5 | 3.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.4 | 1.7 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 3.0 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 4.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.2 | 2.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 1.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 9.6 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 1.9 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 3.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 17.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 2.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 4.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 4.8 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 5.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 4.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.7 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.9 | 49.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.7 | 2.0 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.6 | 1.8 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.5 | 5.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.4 | 4.9 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 3.8 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.3 | 1.5 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.3 | 4.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.3 | 45.0 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 1.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 2.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.2 | 1.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.6 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 1.1 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 2.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.7 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 1.6 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 1.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 16.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 3.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 3.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 6.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 6.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 18.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 3.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 2.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 18.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.1 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 4.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 4.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 1.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.2 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 2.0 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 3.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |