Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
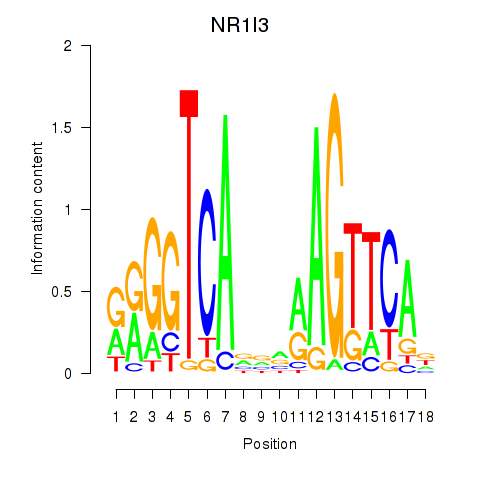
Results for NR1I3
Z-value: 0.14
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg19_v2_chr1_-_161207986_161208000 | -0.38 | 4.9e-09 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_145516560 | 6.20 |
ENST00000537888.1
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr1_+_145516252 | 5.93 |
ENST00000369306.3
|
PEX11B
|
peroxisomal biogenesis factor 11 beta |
| chr14_+_53173910 | 5.77 |
ENST00000606149.1
ENST00000555339.1 ENST00000556813.1 |
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr6_+_32407619 | 5.74 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr14_+_53173890 | 5.73 |
ENST00000445930.2
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr5_+_52856456 | 5.45 |
ENST00000296684.5
ENST00000506765.1 |
NDUFS4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) |
| chr4_-_57524061 | 4.83 |
ENST00000508121.1
|
HOPX
|
HOP homeobox |
| chr5_+_156712372 | 4.49 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chrX_+_118602363 | 4.45 |
ENST00000317881.8
|
SLC25A5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr17_-_62502399 | 4.23 |
ENST00000450599.2
ENST00000585060.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_-_225616515 | 4.10 |
ENST00000338179.2
ENST00000425080.1 |
LBR
|
lamin B receptor |
| chr11_-_82611448 | 4.04 |
ENST00000393399.2
ENST00000313010.3 |
PRCP
|
prolylcarboxypeptidase (angiotensinase C) |
| chr2_-_209118974 | 3.90 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr12_-_118810688 | 3.86 |
ENST00000542532.1
ENST00000392533.3 |
TAOK3
|
TAO kinase 3 |
| chr1_+_153330322 | 3.76 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr16_-_66968055 | 3.73 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr17_-_62502639 | 3.68 |
ENST00000225792.5
ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr19_-_48894104 | 3.59 |
ENST00000597017.1
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr1_-_149900122 | 3.35 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr16_-_66968265 | 3.25 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr10_-_73848764 | 2.73 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr2_-_86790593 | 2.72 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr17_-_53499310 | 2.71 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr18_-_19284724 | 2.67 |
ENST00000580981.1
ENST00000289119.2 |
ABHD3
|
abhydrolase domain containing 3 |
| chr14_-_22005018 | 2.62 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_-_31509714 | 2.35 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr3_-_58419537 | 2.28 |
ENST00000474765.1
ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB
|
pyruvate dehydrogenase (lipoamide) beta |
| chr7_+_143079000 | 2.20 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr11_-_59950519 | 2.16 |
ENST00000528851.1
|
MS4A6A
|
membrane-spanning 4-domains, subfamily A, member 6A |
| chr9_-_75567962 | 2.02 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr15_+_85523671 | 1.94 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr12_+_53693812 | 1.92 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr16_+_67906919 | 1.85 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr12_+_100594557 | 1.82 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr8_+_11351876 | 1.81 |
ENST00000529894.1
|
BLK
|
B lymphoid tyrosine kinase |
| chr1_-_205290865 | 1.71 |
ENST00000367157.3
|
NUAK2
|
NUAK family, SNF1-like kinase, 2 |
| chr7_+_101460882 | 1.60 |
ENST00000292535.7
ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1
|
cut-like homeobox 1 |
| chr15_+_43985084 | 1.57 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr1_-_23886285 | 1.53 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr19_-_6604094 | 1.46 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr9_-_35361262 | 1.41 |
ENST00000599954.1
|
AL160274.1
|
HCG17281; PRO0038; Uncharacterized protein |
| chr15_+_43885252 | 1.41 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr17_-_26695013 | 1.39 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr8_-_53626974 | 1.38 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chrX_-_110655391 | 1.37 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr17_-_26694979 | 1.35 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr1_-_1690014 | 1.30 |
ENST00000400922.2
ENST00000342348.5 |
NADK
|
NAD kinase |
| chr1_+_26036093 | 1.18 |
ENST00000374329.1
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr18_-_19283649 | 1.15 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr1_-_32210275 | 1.06 |
ENST00000440175.2
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr4_+_70894130 | 0.77 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr18_-_23671139 | 0.72 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr16_-_4465886 | 0.64 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr9_-_80646374 | 0.54 |
ENST00000286548.4
|
GNAQ
|
guanine nucleotide binding protein (G protein), q polypeptide |
| chr13_-_95131923 | 0.51 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr14_-_22005062 | 0.37 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr1_+_32930647 | 0.21 |
ENST00000609129.1
|
ZBTB8B
|
zinc finger and BTB domain containing 8B |
| chr1_+_154377669 | 0.17 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr18_-_23670546 | 0.14 |
ENST00000542743.1
ENST00000545952.1 ENST00000539849.1 ENST00000415083.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr9_+_35605274 | 0.12 |
ENST00000336395.5
|
TESK1
|
testis-specific kinase 1 |
| chr6_+_96969672 | 0.09 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr5_-_39270725 | 0.05 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr1_+_44401479 | 0.04 |
ENST00000438616.3
|
ARTN
|
artemin |
| chr19_+_676385 | 0.03 |
ENST00000166139.4
|
FSTL3
|
follistatin-like 3 (secreted glycoprotein) |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.4 | 5.7 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 1.3 | 3.9 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 1.3 | 3.8 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.0 | 4.0 | GO:0002254 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.7 | 4.5 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.7 | 2.0 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 4.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.5 | 7.9 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.5 | 11.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 1.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 2.7 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.3 | 3.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 7.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 2.4 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.3 | 1.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 2.3 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 1.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 4.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.2 | 1.2 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.2 | 3.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 3.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 2.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.5 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 5.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 3.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.9 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 0.5 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.1 | 1.3 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.1 | 1.8 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 1.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 4.1 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.8 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 3.0 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 2.2 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.4 | GO:0071817 | MMXD complex(GO:0071817) |
| 1.0 | 11.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.5 | 12.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 7.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.3 | 1.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.3 | 5.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 3.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.2 | 1.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 2.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 2.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 2.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 4.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 3.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 3.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 5.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 2.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 2.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.3 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 1.3 | 7.9 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 1.3 | 3.9 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 1.3 | 3.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 1.1 | 4.5 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 1.0 | 11.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.9 | 3.6 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.8 | 3.8 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.6 | 2.3 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.4 | 5.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.4 | 2.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.3 | 2.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 4.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 5.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 3.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 2.7 | GO:0004672 | protein kinase activity(GO:0004672) |
| 0.1 | 0.2 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.4 | GO:0032947 | protein complex scaffold(GO:0032947) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 6.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 7.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 4.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 11.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 2.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 4.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 4.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 5.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |