Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
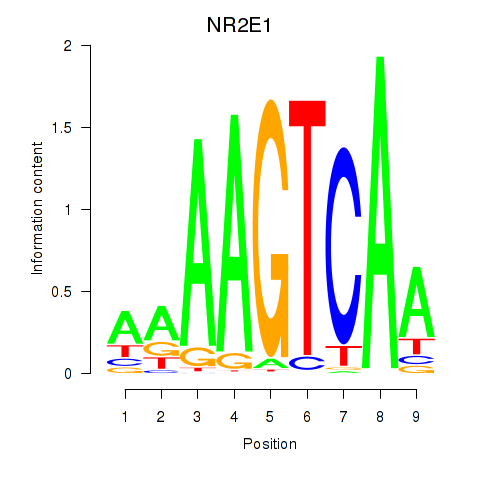
Results for NR2E1
Z-value: 0.68
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | 0.39 | 1.7e-09 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_21786179 | 60.50 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr12_-_91539918 | 49.80 |
ENST00000548218.1
|
DCN
|
decorin |
| chr11_-_111794446 | 26.79 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr5_-_42811986 | 25.70 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr12_-_91572278 | 20.95 |
ENST00000425043.1
ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN
|
decorin |
| chr11_-_5276008 | 19.93 |
ENST00000336906.4
|
HBG2
|
hemoglobin, gamma G |
| chr11_-_5271122 | 19.65 |
ENST00000330597.3
|
HBG1
|
hemoglobin, gamma A |
| chr14_+_69865401 | 16.78 |
ENST00000556605.1
ENST00000336643.5 ENST00000031146.4 |
SLC39A9
|
solute carrier family 39, member 9 |
| chr11_-_111781454 | 15.87 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
| chr17_-_66951474 | 14.95 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr12_-_91546926 | 14.46 |
ENST00000550758.1
|
DCN
|
decorin |
| chr11_-_111781554 | 12.51 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr17_-_26695013 | 11.47 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr1_+_25944341 | 11.27 |
ENST00000263979.3
|
MAN1C1
|
mannosidase, alpha, class 1C, member 1 |
| chr6_+_31916733 | 10.56 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr17_-_26694979 | 10.37 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr17_-_42327236 | 10.34 |
ENST00000399246.2
|
AC003102.1
|
AC003102.1 |
| chr17_-_46667594 | 10.02 |
ENST00000476342.1
ENST00000460160.1 ENST00000472863.1 |
HOXB3
|
homeobox B3 |
| chr8_+_80523962 | 10.01 |
ENST00000518491.1
|
STMN2
|
stathmin-like 2 |
| chr5_-_160279207 | 9.72 |
ENST00000327245.5
|
ATP10B
|
ATPase, class V, type 10B |
| chr17_-_46667628 | 9.60 |
ENST00000498678.1
|
HOXB3
|
homeobox B3 |
| chr1_+_150229554 | 9.60 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr7_+_142829162 | 9.48 |
ENST00000291009.3
|
PIP
|
prolactin-induced protein |
| chr3_+_52812523 | 9.38 |
ENST00000540715.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr1_+_152850787 | 9.35 |
ENST00000368765.3
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein |
| chr12_+_10366016 | 8.83 |
ENST00000546017.1
ENST00000535576.1 ENST00000539170.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr8_-_86253888 | 8.81 |
ENST00000522389.1
ENST00000432364.2 ENST00000517618.1 |
CA1
|
carbonic anhydrase I |
| chr17_-_29624343 | 8.79 |
ENST00000247271.4
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr17_-_43138463 | 8.38 |
ENST00000310604.4
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr7_-_100026280 | 8.36 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr6_+_161123270 | 8.25 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_-_46922659 | 8.25 |
ENST00000265417.7
|
GPR116
|
G protein-coupled receptor 116 |
| chr10_+_124320156 | 8.16 |
ENST00000338354.3
ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1
|
deleted in malignant brain tumors 1 |
| chr10_+_11784360 | 8.11 |
ENST00000379215.4
ENST00000420401.1 |
ECHDC3
|
enoyl CoA hydratase domain containing 3 |
| chr14_-_93799360 | 7.96 |
ENST00000334746.5
ENST00000554565.1 ENST00000298896.3 |
BTBD7
|
BTB (POZ) domain containing 7 |
| chr3_+_14989186 | 7.95 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_+_76262552 | 7.71 |
ENST00000263187.3
|
MSH4
|
mutS homolog 4 |
| chr11_-_62783276 | 7.49 |
ENST00000535878.1
ENST00000545207.1 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr11_-_62783303 | 7.44 |
ENST00000336232.2
ENST00000430500.2 |
SLC22A8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr17_+_29664830 | 7.40 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chrX_-_65253506 | 7.38 |
ENST00000427538.1
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr2_+_67624430 | 7.31 |
ENST00000272342.5
|
ETAA1
|
Ewing tumor-associated antigen 1 |
| chr5_-_135290705 | 7.22 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr3_+_52813932 | 7.13 |
ENST00000537050.1
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr10_+_124320195 | 7.10 |
ENST00000359586.6
|
DMBT1
|
deleted in malignant brain tumors 1 |
| chr12_-_52887034 | 7.05 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr3_+_69788576 | 6.85 |
ENST00000352241.4
ENST00000448226.2 |
MITF
|
microphthalmia-associated transcription factor |
| chr5_-_140013275 | 6.67 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr17_+_73975292 | 6.61 |
ENST00000397640.1
ENST00000416485.1 ENST00000588202.1 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 CST complex subunit |
| chr9_-_99064429 | 6.58 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr9_+_87285539 | 6.53 |
ENST00000359847.3
|
NTRK2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr15_-_88799384 | 6.48 |
ENST00000540489.2
ENST00000557856.1 ENST00000558676.1 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr4_+_96012614 | 6.38 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chrX_-_65259900 | 6.35 |
ENST00000412866.2
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_244006528 | 6.35 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chrX_-_53711064 | 6.24 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr5_-_160973649 | 6.22 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr11_-_62476965 | 6.22 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr14_+_22977587 | 6.21 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chr3_+_69915385 | 6.20 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr16_+_3451184 | 6.20 |
ENST00000575752.1
ENST00000571936.1 ENST00000344823.5 |
ZNF174
|
zinc finger protein 174 |
| chrX_-_153236620 | 6.17 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr17_-_41623691 | 6.14 |
ENST00000545954.1
|
ETV4
|
ets variant 4 |
| chr15_+_58702742 | 6.12 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr17_+_56315787 | 6.11 |
ENST00000262290.4
ENST00000421678.2 |
LPO
|
lactoperoxidase |
| chr3_-_101232019 | 6.10 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chrX_+_136648297 | 6.00 |
ENST00000287538.5
|
ZIC3
|
Zic family member 3 |
| chr3_+_69985792 | 5.98 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chrX_-_65259914 | 5.89 |
ENST00000374737.4
ENST00000455586.2 |
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr8_+_11561660 | 5.88 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr17_-_73975444 | 5.80 |
ENST00000293217.5
ENST00000537812.1 |
ACOX1
|
acyl-CoA oxidase 1, palmitoyl |
| chr13_+_25338290 | 5.78 |
ENST00000255324.5
ENST00000381921.1 ENST00000255325.6 |
RNF17
|
ring finger protein 17 |
| chr14_+_61654271 | 5.67 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr9_-_16727978 | 5.52 |
ENST00000418777.1
ENST00000468187.2 |
BNC2
|
basonuclin 2 |
| chr4_+_88571429 | 5.51 |
ENST00000339673.6
ENST00000282479.7 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr11_-_57004658 | 5.27 |
ENST00000606794.1
|
APLNR
|
apelin receptor |
| chr10_-_115423792 | 5.26 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr20_-_4229721 | 5.25 |
ENST00000379453.4
|
ADRA1D
|
adrenoceptor alpha 1D |
| chr12_+_111471828 | 5.20 |
ENST00000261726.6
|
CUX2
|
cut-like homeobox 2 |
| chr10_-_21463116 | 5.17 |
ENST00000417816.2
|
NEBL
|
nebulette |
| chr4_-_47983519 | 5.16 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr9_-_91793675 | 5.11 |
ENST00000375835.4
ENST00000375830.1 |
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr1_+_32042105 | 5.10 |
ENST00000457433.2
ENST00000441210.2 |
TINAGL1
|
tubulointerstitial nephritis antigen-like 1 |
| chrX_-_106362013 | 5.10 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr17_-_41623716 | 5.06 |
ENST00000319349.5
|
ETV4
|
ets variant 4 |
| chr20_-_30310656 | 5.05 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr7_-_8301768 | 4.98 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr6_+_148663729 | 4.92 |
ENST00000367467.3
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_-_116311402 | 4.83 |
ENST00000261448.5
|
CASQ2
|
calsequestrin 2 (cardiac muscle) |
| chr10_-_29084886 | 4.80 |
ENST00000608061.1
ENST00000443246.2 ENST00000446012.1 |
LINC00837
|
long intergenic non-protein coding RNA 837 |
| chr9_-_21368075 | 4.78 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr4_+_71063641 | 4.73 |
ENST00000514097.1
|
ODAM
|
odontogenic, ameloblast asssociated |
| chr9_-_75567962 | 4.72 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr20_-_30310693 | 4.68 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr1_-_48866517 | 4.67 |
ENST00000371841.1
|
SPATA6
|
spermatogenesis associated 6 |
| chr17_-_43138357 | 4.61 |
ENST00000342350.5
|
DCAKD
|
dephospho-CoA kinase domain containing |
| chr2_-_87248975 | 4.59 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr5_+_140743859 | 4.58 |
ENST00000518069.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr16_+_56385290 | 4.58 |
ENST00000564727.1
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr6_+_26156551 | 4.55 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr3_-_116164306 | 4.50 |
ENST00000490035.2
|
LSAMP
|
limbic system-associated membrane protein |
| chr20_+_43160458 | 4.47 |
ENST00000372889.1
ENST00000372887.1 ENST00000372882.3 |
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr2_+_113885138 | 4.44 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr18_+_29027696 | 4.42 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr21_-_36260980 | 4.37 |
ENST00000344691.4
ENST00000358356.5 |
RUNX1
|
runt-related transcription factor 1 |
| chr5_-_9546180 | 4.37 |
ENST00000382496.5
|
SEMA5A
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr6_+_31515337 | 4.35 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr5_-_33892046 | 4.34 |
ENST00000352040.3
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr5_-_11588907 | 4.31 |
ENST00000513598.1
ENST00000503622.1 |
CTNND2
|
catenin (cadherin-associated protein), delta 2 |
| chr2_+_88047606 | 4.28 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr5_-_33892204 | 4.27 |
ENST00000504830.1
|
ADAMTS12
|
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
| chr7_-_8301682 | 4.27 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chrX_-_33146477 | 4.23 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr2_-_196933536 | 4.23 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr20_+_57875457 | 4.14 |
ENST00000337938.2
ENST00000311585.7 ENST00000371028.2 |
EDN3
|
endothelin 3 |
| chrX_+_47078069 | 4.10 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr1_-_20834586 | 4.05 |
ENST00000264198.3
|
MUL1
|
mitochondrial E3 ubiquitin protein ligase 1 |
| chr7_-_113559104 | 3.94 |
ENST00000284601.3
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A |
| chr12_-_71031220 | 3.88 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chrX_+_150866779 | 3.68 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr16_-_66952742 | 3.68 |
ENST00000565235.2
ENST00000568632.1 ENST00000565796.1 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr12_-_71031185 | 3.63 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr16_+_47496023 | 3.57 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr1_-_154155595 | 3.54 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chr3_-_148939835 | 3.52 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr5_+_137203465 | 3.47 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr4_+_3388057 | 3.43 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr6_-_161085291 | 3.42 |
ENST00000316300.5
|
LPA
|
lipoprotein, Lp(a) |
| chr2_+_3705785 | 3.42 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr9_-_5304432 | 3.39 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr10_+_17794251 | 3.38 |
ENST00000377495.1
ENST00000338221.5 |
TMEM236
|
transmembrane protein 236 |
| chr3_+_113616317 | 3.35 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr18_+_22040593 | 3.34 |
ENST00000256906.4
|
HRH4
|
histamine receptor H4 |
| chr3_+_42897512 | 3.32 |
ENST00000493193.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr6_-_41715128 | 3.31 |
ENST00000356667.4
ENST00000373025.3 ENST00000425343.2 |
PGC
|
progastricsin (pepsinogen C) |
| chr14_+_31494841 | 3.30 |
ENST00000556232.1
ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1
|
adaptor-related protein complex 4, sigma 1 subunit |
| chr11_-_30038490 | 3.22 |
ENST00000328224.6
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr3_+_189349162 | 3.13 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr12_-_122018859 | 3.11 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr7_-_107443652 | 2.98 |
ENST00000340010.5
ENST00000422236.2 ENST00000453332.1 |
SLC26A3
|
solute carrier family 26 (anion exchanger), member 3 |
| chr4_+_4861385 | 2.96 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr4_-_123542224 | 2.94 |
ENST00000264497.3
|
IL21
|
interleukin 21 |
| chr2_-_169887827 | 2.90 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr9_-_95166841 | 2.89 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr6_+_106546808 | 2.89 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr20_-_30310797 | 2.88 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr5_-_127873496 | 2.87 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr7_+_39125365 | 2.85 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr12_-_58135903 | 2.82 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr13_+_109248500 | 2.81 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr2_+_11679963 | 2.77 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr5_+_32711829 | 2.70 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chrX_+_47444613 | 2.68 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr2_-_74555293 | 2.68 |
ENST00000358683.4
|
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr5_-_55412774 | 2.67 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr3_-_58196939 | 2.67 |
ENST00000394549.2
ENST00000461914.3 |
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr22_-_40929812 | 2.65 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr17_+_1959369 | 2.62 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
hypermethylated in cancer 1 |
| chr1_+_159141397 | 2.61 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr18_+_22040620 | 2.60 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr9_-_5339873 | 2.59 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr6_+_131521120 | 2.53 |
ENST00000537868.1
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr12_-_56236690 | 2.52 |
ENST00000322569.4
|
MMP19
|
matrix metallopeptidase 19 |
| chr16_-_10652993 | 2.51 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2 |
| chr7_-_151217001 | 2.51 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr4_-_141348999 | 2.50 |
ENST00000325617.5
|
CLGN
|
calmegin |
| chr15_-_42783303 | 2.50 |
ENST00000565380.1
ENST00000564754.1 |
ZNF106
|
zinc finger protein 106 |
| chr11_-_13517565 | 2.46 |
ENST00000282091.1
ENST00000529816.1 |
PTH
|
parathyroid hormone |
| chr2_-_183291741 | 2.44 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr6_+_33048222 | 2.43 |
ENST00000428835.1
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1 |
| chr1_-_154155675 | 2.43 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr8_+_77593448 | 2.43 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr3_+_69985734 | 2.42 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr4_-_70505358 | 2.38 |
ENST00000457664.2
ENST00000604629.1 ENST00000604021.1 |
UGT2A2
|
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr3_-_58196688 | 2.37 |
ENST00000486455.1
|
DNASE1L3
|
deoxyribonuclease I-like 3 |
| chr1_-_68698222 | 2.34 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr7_+_134832808 | 2.27 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr19_-_633576 | 2.27 |
ENST00000588649.2
|
POLRMT
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr16_-_73082274 | 2.25 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr12_-_82152444 | 2.22 |
ENST00000549325.1
ENST00000550584.2 |
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr11_-_83984231 | 2.21 |
ENST00000330014.6
ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr11_-_8832182 | 2.20 |
ENST00000527510.1
ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5
|
suppression of tumorigenicity 5 |
| chr3_+_46616017 | 2.20 |
ENST00000542931.1
|
TDGF1
|
teratocarcinoma-derived growth factor 1 |
| chr4_-_122085469 | 2.19 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr5_-_127873659 | 2.19 |
ENST00000262464.4
|
FBN2
|
fibrillin 2 |
| chr9_+_21440440 | 2.16 |
ENST00000276927.1
|
IFNA1
|
interferon, alpha 1 |
| chr3_+_148508845 | 2.12 |
ENST00000491148.1
|
CPB1
|
carboxypeptidase B1 (tissue) |
| chr16_-_20709066 | 2.12 |
ENST00000520010.1
|
ACSM1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr7_-_14942283 | 2.08 |
ENST00000402815.1
|
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr16_-_66952779 | 2.08 |
ENST00000570262.1
ENST00000394055.3 ENST00000299752.4 |
CDH16
|
cadherin 16, KSP-cadherin |
| chr11_+_63742050 | 2.03 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr9_-_95166884 | 2.01 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr1_-_154150651 | 2.01 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr5_+_137203557 | 2.00 |
ENST00000515645.1
|
MYOT
|
myotilin |
| chr7_+_80267973 | 1.97 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr17_+_19186292 | 1.97 |
ENST00000395626.1
ENST00000571254.1 |
EPN2
|
epsin 2 |
| chr2_+_90077680 | 1.96 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr15_-_43559055 | 1.96 |
ENST00000220420.5
ENST00000349114.4 |
TGM5
|
transglutaminase 5 |
| chr17_-_41738931 | 1.91 |
ENST00000329168.3
ENST00000549132.1 |
MEOX1
|
mesenchyme homeobox 1 |
| chr7_-_72936608 | 1.91 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 85.8 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 4.3 | 8.6 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902548) |
| 3.9 | 19.6 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 3.1 | 55.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 2.7 | 8.2 | GO:2000048 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.6 | 39.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 2.5 | 10.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 2.5 | 7.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 2.4 | 7.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 2.2 | 6.5 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 2.0 | 5.9 | GO:0003285 | septum secundum development(GO:0003285) |
| 1.9 | 11.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 1.8 | 1.8 | GO:0050880 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 1.7 | 6.7 | GO:0038123 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 1.6 | 4.7 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 1.6 | 4.7 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.5 | 6.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 1.5 | 15.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.5 | 11.9 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 1.4 | 8.3 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 1.4 | 15.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.3 | 9.4 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 1.3 | 5.2 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 1.3 | 6.5 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 1.3 | 6.4 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.2 | 4.9 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 1.2 | 1.2 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 1.2 | 30.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.2 | 10.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.1 | 3.4 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 1.0 | 3.1 | GO:0021678 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 1.0 | 4.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.0 | 6.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 1.0 | 3.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 1.0 | 2.9 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.9 | 7.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.8 | 2.5 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.8 | 4.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.8 | 3.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.7 | 2.9 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.7 | 5.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.7 | 2.1 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.7 | 2.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.7 | 16.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.6 | 7.1 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.6 | 13.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.6 | 4.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.6 | 15.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.6 | 2.9 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.6 | 1.7 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.6 | 1.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.6 | 4.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.6 | 2.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.6 | 1.7 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.6 | 2.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.6 | 5.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.6 | 1.7 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.6 | 4.4 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.6 | 3.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.5 | 2.7 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.5 | 4.8 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.5 | 5.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.5 | 8.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 1.9 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.5 | 9.5 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.5 | 1.9 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 1.8 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 1.3 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.4 | 2.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.4 | 3.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.4 | 21.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 21.7 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.4 | 4.8 | GO:0086068 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.4 | 5.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 4.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 9.7 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.3 | 7.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 14.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 5.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 4.4 | GO:0048843 | positive regulation of axon extension involved in axon guidance(GO:0048842) negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.3 | 2.7 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.3 | 2.0 | GO:0071724 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.3 | 6.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 4.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 1.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.3 | 2.7 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.3 | 4.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 4.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 | 4.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.3 | 5.0 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.3 | 4.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 5.5 | GO:0043586 | tongue development(GO:0043586) |
| 0.3 | 6.6 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.3 | 4.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.3 | 6.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 5.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 4.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.2 | 0.9 | GO:0048840 | otolith development(GO:0048840) |
| 0.2 | 2.2 | GO:0007350 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.2 | 2.6 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 5.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 2.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 4.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.2 | 1.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 2.5 | GO:2001212 | regulation of vasculogenesis(GO:2001212) |
| 0.1 | 0.7 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 5.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 1.7 | GO:0051775 | response to redox state(GO:0051775) |
| 0.1 | 2.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.1 | 4.6 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 4.1 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 2.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 1.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 8.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 2.8 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.1 | 5.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 3.7 | GO:0010863 | positive regulation of phospholipase C activity(GO:0010863) |
| 0.1 | 2.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 2.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.0 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 2.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.8 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 6.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 2.5 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.1 | 0.5 | GO:0042481 | regulation of odontogenesis(GO:0042481) |
| 0.1 | 7.3 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 6.1 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.1 | 1.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 7.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.0 | GO:0006897 | endocytosis(GO:0006897) |
| 0.1 | 2.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 2.4 | GO:0032729 | positive regulation of interferon-gamma production(GO:0032729) |
| 0.0 | 2.8 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 2.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 1.0 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 4.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 3.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 9.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 2.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 3.4 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 1.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) regulation of melanin biosynthetic process(GO:0048021) positive regulation of melanin biosynthetic process(GO:0048023) regulation of secondary metabolite biosynthetic process(GO:1900376) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 3.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 2.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 1.1 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0002525 | acute inflammatory response to non-antigenic stimulus(GO:0002525) regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.3 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 4.8 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 1.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:0042109 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 1.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 85.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 3.4 | 55.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.8 | 39.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.6 | 10.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.9 | 7.7 | GO:0005713 | recombination nodule(GO:0005713) |
| 1.4 | 8.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.3 | 6.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.2 | 4.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.2 | 4.7 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.9 | 15.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.9 | 5.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.8 | 10.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.5 | 6.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.5 | 6.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.5 | 4.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.4 | 8.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 2.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 7.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 6.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.3 | 1.7 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.3 | 4.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 5.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 4.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.3 | 11.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 8.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 5.1 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 5.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 6.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 7.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 4.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 1.9 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.1 | 11.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 2.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 9.5 | GO:0034358 | plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.1 | 4.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 2.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 5.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.1 | 17.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 5.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 4.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.0 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.1 | 16.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 2.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 10.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 2.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 10.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 7.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 4.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 4.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 4.9 | GO:0043234 | protein complex(GO:0043234) |
| 0.0 | 4.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 6.2 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 3.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 4.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 48.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 5.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 4.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 2.3 | GO:0000428 | DNA-directed RNA polymerase complex(GO:0000428) RNA polymerase complex(GO:0030880) |
| 0.0 | 1.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.3 | GO:0043235 | receptor complex(GO:0043235) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 13.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 3.0 | 39.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 2.2 | 6.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 1.9 | 15.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.8 | 57.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.7 | 5.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 1.5 | 4.4 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 1.4 | 8.3 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 1.3 | 6.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 1.3 | 95.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 1.1 | 5.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.1 | 8.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 1.1 | 4.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.0 | 8.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 1.0 | 11.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.9 | 4.7 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.9 | 10.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.9 | 6.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.8 | 6.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.8 | 4.5 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.7 | 2.9 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.7 | 3.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 17.2 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.7 | 6.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.7 | 9.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 5.9 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.6 | 5.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.6 | 20.6 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.6 | 5.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.6 | 8.8 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.6 | 12.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.6 | 7.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.6 | 5.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.5 | 5.9 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.5 | 6.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.5 | 6.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.5 | 2.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.4 | 9.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 1.6 | GO:0033906 | protein tyrosine kinase inhibitor activity(GO:0030292) hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 5.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.4 | 2.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 3.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 4.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.4 | 3.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.4 | 7.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.4 | 6.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.4 | 21.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 1.8 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.3 | 1.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 3.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 10.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.3 | 3.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 7.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 12.2 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 3.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 4.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 4.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 1.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 5.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 2.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 3.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 2.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.2 | 1.9 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.2 | 4.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 0.8 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 4.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.2 | 4.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 19.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.2 | 7.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.2 | 1.3 | GO:0005497 | androgen binding(GO:0005497) |
| 0.2 | 1.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 16.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 3.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.2 | 2.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 2.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 14.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 10.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 2.5 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 8.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 6.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 4.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.0 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 2.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 2.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 9.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 2.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 7.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 2.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 2.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.4 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 3.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 2.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 5.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 2.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 1.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 8.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 10.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 2.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.8 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 4.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 82.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 10.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.4 | 18.6 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.3 | 10.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 6.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.3 | 4.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 28.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 4.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 7.9 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 12.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 1.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.2 | 6.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 9.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 9.3 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 5.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 6.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 35.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 11.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 29.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 7.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 2.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 6.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 1.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 6.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.2 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 2.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 12.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 85.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 1.2 | 18.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.9 | 5.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.7 | 14.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.6 | 8.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.6 | 5.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.6 | 4.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 6.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.5 | 10.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 15.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.4 | 4.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 10.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.3 | 5.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.3 | 5.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 8.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 4.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.3 | 6.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 1.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 1.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.2 | 10.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 12.8 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.2 | 6.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.4 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.2 | 1.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 4.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 9.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 7.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 12.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 6.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 2.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 4.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 15.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 3.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.8 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 3.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 2.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |