Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
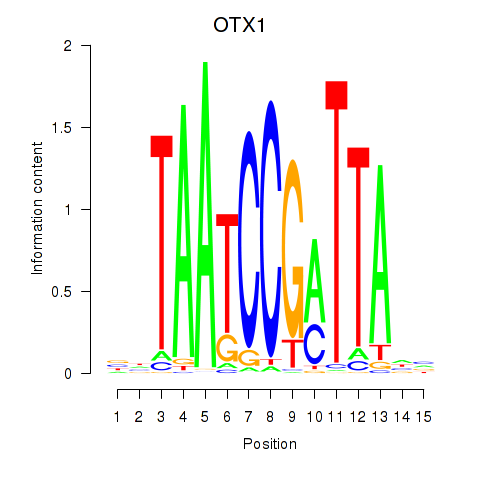
Results for OTX1
Z-value: 1.35
Transcription factors associated with OTX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX1
|
ENSG00000115507.5 | orthodenticle homeobox 1 |
Activity profile of OTX1 motif
Sorted Z-values of OTX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_177631497 | 31.60 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr11_-_65626753 | 28.78 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr12_-_54071181 | 25.26 |
ENST00000338662.5
|
ATP5G2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) |
| chr16_-_58163299 | 24.04 |
ENST00000262498.3
|
C16orf80
|
chromosome 16 open reading frame 80 |
| chr16_+_33605231 | 23.59 |
ENST00000570121.2
|
IGHV3OR16-12
|
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr2_-_85637459 | 22.09 |
ENST00000409921.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr2_-_161350305 | 22.02 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr4_-_122744998 | 19.92 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr3_+_138340067 | 19.35 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_-_47616992 | 18.85 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr3_+_138340049 | 18.00 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_-_106054952 | 16.84 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr2_+_201936707 | 16.83 |
ENST00000433898.1
ENST00000454214.1 |
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr20_-_33539655 | 16.29 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr9_-_116172946 | 15.69 |
ENST00000374171.4
|
POLE3
|
polymerase (DNA directed), epsilon 3, accessory subunit |
| chr1_-_36930012 | 15.58 |
ENST00000373116.5
|
MRPS15
|
mitochondrial ribosomal protein S15 |
| chr14_-_102552659 | 15.06 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chrX_-_109561294 | 14.53 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr1_+_40723779 | 14.49 |
ENST00000372759.3
|
ZMPSTE24
|
zinc metallopeptidase STE24 |
| chr3_+_142315225 | 14.28 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr17_+_41158742 | 14.11 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr1_-_205719295 | 13.71 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr2_+_122513109 | 13.42 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr19_-_10530784 | 13.31 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr5_+_135496675 | 12.98 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chr2_+_201936458 | 12.88 |
ENST00000237889.4
|
NDUFB3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr16_-_69373396 | 12.86 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr5_-_77072085 | 12.84 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr6_-_41039567 | 12.14 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chrX_+_100645812 | 12.10 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr2_-_74619152 | 12.08 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr18_+_8717369 | 11.96 |
ENST00000359865.3
ENST00000400050.3 ENST00000306285.7 |
SOGA2
|
SOGA family member 2 |
| chr3_-_69129501 | 11.86 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr6_-_144416737 | 11.79 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr17_+_56769924 | 11.58 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr8_+_101162812 | 11.27 |
ENST00000353107.3
ENST00000522439.1 |
POLR2K
|
polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa |
| chr22_+_30163340 | 11.01 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr16_-_29875057 | 10.95 |
ENST00000219789.6
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chrX_-_134156502 | 10.68 |
ENST00000391440.1
|
FAM127C
|
family with sequence similarity 127, member C |
| chr6_-_34393825 | 10.27 |
ENST00000605528.1
ENST00000326199.8 |
RPS10-NUDT3
RPS10
|
RPS10-NUDT3 readthrough ribosomal protein S10 |
| chr12_+_20963632 | 9.99 |
ENST00000540853.1
ENST00000261196.2 |
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr5_+_96038476 | 9.92 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr12_+_20963647 | 9.76 |
ENST00000381545.3
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3 |
| chr20_-_32700075 | 9.44 |
ENST00000374980.2
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
| chr8_+_132916318 | 9.18 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr3_-_141747950 | 9.16 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr11_-_104817919 | 9.14 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr1_+_174844645 | 9.08 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr11_-_67275542 | 8.89 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr3_+_136649311 | 8.85 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr4_+_83956237 | 8.82 |
ENST00000264389.2
|
COPS4
|
COP9 signalosome subunit 4 |
| chr4_+_83956312 | 8.72 |
ENST00000509317.1
ENST00000503682.1 ENST00000511653.1 |
COPS4
|
COP9 signalosome subunit 4 |
| chr11_-_67276100 | 8.52 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr12_-_14956396 | 8.49 |
ENST00000535328.1
ENST00000261167.2 |
WBP11
|
WW domain binding protein 11 |
| chr1_+_74701062 | 8.44 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr16_-_29874462 | 8.35 |
ENST00000566113.1
ENST00000569956.1 ENST00000570016.1 |
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr12_+_6603253 | 7.61 |
ENST00000382457.4
ENST00000545962.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chr14_+_21458127 | 7.58 |
ENST00000382985.4
ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17
|
methyltransferase like 17 |
| chr5_-_111091948 | 7.46 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr8_-_145652336 | 7.37 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr12_+_57881740 | 7.26 |
ENST00000262027.5
ENST00000315473.5 |
MARS
|
methionyl-tRNA synthetase |
| chr6_+_150070857 | 7.20 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr1_+_81771806 | 7.20 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr1_-_9811600 | 6.94 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
| chr11_-_67210930 | 6.65 |
ENST00000453768.2
ENST00000545016.1 ENST00000341356.5 |
CORO1B
|
coronin, actin binding protein, 1B |
| chr5_-_54603368 | 6.63 |
ENST00000508346.1
ENST00000251636.5 |
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr2_-_74618964 | 6.55 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chrX_+_149861836 | 6.45 |
ENST00000542156.1
ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1
|
myotubularin related protein 1 |
| chr5_-_81574160 | 6.37 |
ENST00000510210.1
ENST00000512493.1 ENST00000507980.1 ENST00000511844.1 ENST00000510019.1 |
RPS23
|
ribosomal protein S23 |
| chr1_+_150293921 | 6.33 |
ENST00000324862.6
|
PRPF3
|
pre-mRNA processing factor 3 |
| chr1_+_109756523 | 6.10 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr1_-_89458604 | 6.05 |
ENST00000260508.4
|
CCBL2
|
cysteine conjugate-beta lyase 2 |
| chr15_-_43876702 | 5.94 |
ENST00000348806.6
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr7_-_100895414 | 5.69 |
ENST00000435848.1
ENST00000474120.1 |
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chr1_+_172502336 | 5.68 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr1_+_150293973 | 5.62 |
ENST00000414970.2
ENST00000543398.1 |
PRPF3
|
pre-mRNA processing factor 3 |
| chr8_+_109455845 | 5.48 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr10_+_60028818 | 5.36 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr19_+_21265028 | 5.26 |
ENST00000291770.7
|
ZNF714
|
zinc finger protein 714 |
| chr2_-_112642267 | 5.17 |
ENST00000341068.3
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr16_-_8962200 | 5.05 |
ENST00000562843.1
ENST00000561530.1 ENST00000396593.2 |
CARHSP1
|
calcium regulated heat stable protein 1, 24kDa |
| chr11_-_111957451 | 4.87 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr7_+_128379346 | 4.87 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr12_+_100594557 | 4.82 |
ENST00000546902.1
ENST00000552376.1 ENST00000551617.1 |
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast) |
| chr11_-_65626797 | 4.82 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr11_-_78052923 | 4.78 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr9_+_125132803 | 4.73 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_83769996 | 4.51 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr16_-_29874211 | 4.36 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr14_-_106539557 | 4.31 |
ENST00000390599.2
|
IGHV1-8
|
immunoglobulin heavy variable 1-8 |
| chr14_+_50234309 | 4.22 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr9_+_128509624 | 4.09 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr1_-_216596738 | 3.92 |
ENST00000307340.3
ENST00000366943.2 ENST00000366942.3 |
USH2A
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr6_+_26251835 | 3.81 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr14_-_102706934 | 3.75 |
ENST00000523231.1
ENST00000524370.1 ENST00000517966.1 |
MOK
|
MOK protein kinase |
| chr7_+_128379449 | 3.73 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr19_-_41903161 | 3.69 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr8_-_56986768 | 3.65 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr2_+_204103663 | 3.38 |
ENST00000356079.4
ENST00000429815.2 |
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr16_-_21868978 | 3.29 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr15_+_42787452 | 3.21 |
ENST00000249647.3
|
SNAP23
|
synaptosomal-associated protein, 23kDa |
| chr10_-_35104185 | 3.20 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr14_-_107170409 | 3.17 |
ENST00000390633.2
|
IGHV1-69
|
immunoglobulin heavy variable 1-69 |
| chr2_-_47403642 | 2.90 |
ENST00000456319.1
ENST00000409563.1 ENST00000272298.7 |
CALM2
|
calmodulin 2 (phosphorylase kinase, delta) |
| chr5_-_39425222 | 2.80 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr16_+_14802801 | 2.56 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr6_-_137539651 | 2.51 |
ENST00000543628.1
|
IFNGR1
|
interferon gamma receptor 1 |
| chr6_+_26158343 | 2.47 |
ENST00000377777.4
ENST00000289316.2 |
HIST1H2BD
|
histone cluster 1, H2bd |
| chrX_-_148571884 | 2.38 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr14_-_24911448 | 2.37 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr12_+_113354341 | 2.35 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_28764653 | 2.35 |
ENST00000373836.3
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr16_-_21436459 | 2.34 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr14_-_106471723 | 2.28 |
ENST00000390595.2
|
IGHV1-3
|
immunoglobulin heavy variable 1-3 |
| chr13_+_38923959 | 2.16 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr11_-_62420757 | 2.15 |
ENST00000330574.2
|
INTS5
|
integrator complex subunit 5 |
| chr14_-_107211459 | 2.10 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr7_-_100239132 | 2.06 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr18_+_616672 | 2.05 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_+_8704748 | 1.98 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr5_-_39425290 | 1.96 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr17_-_26876350 | 1.88 |
ENST00000470125.1
|
UNC119
|
unc-119 homolog (C. elegans) |
| chr16_+_69373323 | 1.86 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr3_-_45017609 | 1.85 |
ENST00000342790.4
ENST00000424952.2 ENST00000296127.3 ENST00000455235.1 |
ZDHHC3
|
zinc finger, DHHC-type containing 3 |
| chr2_-_49381646 | 1.78 |
ENST00000346173.3
ENST00000406846.2 |
FSHR
|
follicle stimulating hormone receptor |
| chr1_-_186430222 | 1.77 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr16_-_29415350 | 1.76 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr19_-_42916499 | 1.65 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr19_+_35940486 | 1.64 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr4_-_70626314 | 1.42 |
ENST00000510821.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr17_-_36981556 | 1.34 |
ENST00000536127.1
ENST00000225428.5 |
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr8_-_33370607 | 1.32 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr3_+_11314099 | 1.30 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr22_+_39052632 | 1.10 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr2_+_232135245 | 1.06 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr13_-_31736478 | 0.92 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr19_-_3761673 | 0.71 |
ENST00000316757.3
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr18_+_616711 | 0.68 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chrX_-_43832711 | 0.50 |
ENST00000378062.5
|
NDP
|
Norrie disease (pseudoglioma) |
| chr15_-_61521495 | 0.33 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr8_-_79717750 | 0.27 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr1_-_151300160 | 0.21 |
ENST00000368874.4
|
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr20_+_48884002 | 0.13 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr6_+_96025341 | 0.07 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr2_+_1418154 | 0.02 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.7 | 33.6 | GO:2000812 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 5.0 | 19.9 | GO:0071373 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 4.8 | 14.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 4.0 | 11.9 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 3.8 | 15.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 3.5 | 17.4 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 3.4 | 13.7 | GO:0019046 | release from viral latency(GO:0019046) |
| 3.4 | 16.8 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 3.0 | 8.9 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 2.9 | 14.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.5 | 7.4 | GO:2000395 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 2.3 | 18.6 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 2.3 | 11.6 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 2.2 | 6.7 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 2.2 | 13.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 2.1 | 12.8 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 2.0 | 6.1 | GO:0097056 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 1.4 | 11.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 1.3 | 9.4 | GO:0036093 | male germ cell proliferation(GO:0002176) germ cell proliferation(GO:0036093) |
| 1.3 | 23.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 1.3 | 1.3 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 1.2 | 3.7 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 1.2 | 4.8 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 1.1 | 5.7 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 1.1 | 17.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 1.1 | 22.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 1.0 | 13.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 1.0 | 9.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.0 | 3.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.0 | 7.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.9 | 1.8 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.8 | 7.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.8 | 25.3 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.8 | 2.4 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.7 | 11.9 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.7 | 16.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.7 | 9.9 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.7 | 4.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.6 | 7.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.6 | 13.3 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.5 | 1.6 | GO:0002752 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.5 | 15.7 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.5 | 1.9 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.5 | 19.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.4 | 1.8 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.4 | 28.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.4 | 10.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.4 | 8.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 27.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.4 | 8.4 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.3 | 2.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 5.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 1.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.3 | 11.3 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.3 | 5.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 2.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 0.9 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.3 | 4.8 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.3 | 4.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.3 | 5.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.3 | 4.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 6.4 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.3 | 3.2 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.2 | 3.2 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 12.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.2 | 4.9 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 4.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 15.6 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.2 | 9.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 6.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.2 | 17.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 16.5 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.5 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 1.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 22.0 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.1 | 5.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.5 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 8.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 2.4 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 3.0 | GO:0006412 | translation(GO:0006412) |
| 0.1 | 6.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 20.6 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.9 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 6.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 6.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 12.1 | GO:0042278 | purine nucleoside metabolic process(GO:0042278) |
| 0.0 | 5.2 | GO:0090002 | establishment of protein localization to plasma membrane(GO:0090002) |
| 0.0 | 29.7 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 2.1 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 2.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.9 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 3.1 | 9.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 3.1 | 15.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 2.4 | 14.3 | GO:1990357 | terminal web(GO:1990357) |
| 1.9 | 11.6 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 1.9 | 7.6 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 1.5 | 9.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 1.5 | 22.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 1.4 | 25.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 1.3 | 3.9 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 1.2 | 18.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 1.1 | 7.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 1.0 | 11.0 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.9 | 12.9 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.9 | 33.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.8 | 8.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 11.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.6 | 16.0 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.6 | 4.9 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.6 | 5.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.6 | 15.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.6 | 13.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.5 | 27.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 4.8 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 8.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.5 | 11.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.5 | 8.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.5 | 11.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.5 | 3.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 28.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 7.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 17.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 16.8 | GO:0031430 | M band(GO:0031430) |
| 0.3 | 20.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 5.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 14.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 5.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 38.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 4.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 2.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 5.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 14.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 6.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 3.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 3.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 7.6 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 1.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 6.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
| 0.0 | 13.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 30.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 13.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 9.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 7.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 4.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 2.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 4.7 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 23.7 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 2.4 | 7.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 2.0 | 6.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.8 | 8.9 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.8 | 17.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 1.7 | 9.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 1.6 | 11.0 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.5 | 15.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.5 | 11.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.4 | 11.6 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 1.2 | 4.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 1.2 | 16.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.9 | 25.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.9 | 13.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.9 | 19.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.9 | 4.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.8 | 6.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.8 | 8.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.7 | 9.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.6 | 11.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.6 | 1.7 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.5 | 18.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.5 | 15.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.5 | 28.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 2.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.5 | 2.5 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.5 | 27.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.5 | 6.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.5 | 6.6 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 1.8 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.4 | 55.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.4 | 23.7 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.4 | 5.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.4 | 1.8 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 2.4 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.3 | 35.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.3 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 12.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 4.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 16.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.2 | 2.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 50.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 1.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 14.5 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.2 | 12.1 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.2 | 7.3 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 9.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 6.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 13.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 25.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 4.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 3.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 4.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 7.4 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 3.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 8.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 4.9 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 3.3 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 24.6 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 7.4 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 33.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.8 | 19.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.7 | 13.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.5 | 11.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.5 | 16.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.5 | 23.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.4 | 42.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.4 | 18.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 13.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 2.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 7.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 9.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 9.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 48.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 1.2 | 19.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.7 | 19.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.7 | 7.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.5 | 13.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.5 | 11.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.5 | 12.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 8.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.5 | 16.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 3.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.4 | 29.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.4 | 13.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 29.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 13.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.3 | 23.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.3 | 4.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 11.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 5.2 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.2 | 22.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.2 | 2.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 4.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 9.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.2 | 16.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 3.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 6.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 2.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 14.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 10.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 16.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 1.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.1 | 2.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 4.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 8.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 4.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 3.2 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |