Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
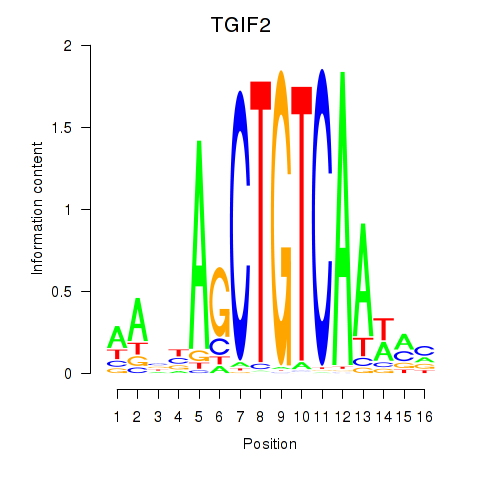
Results for PKNOX1_TGIF2
Z-value: 0.47
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2 | hg19_v2_chr20_+_35202909_35203075 | 0.45 | 2.0e-12 | Click! |
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | -0.19 | 4.2e-03 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_11666649 | 19.59 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr7_-_148580563 | 16.90 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr19_+_49496705 | 15.12 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr1_-_32384693 | 14.54 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_-_63988846 | 13.92 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr18_-_54318353 | 13.77 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr6_+_111195973 | 13.05 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr3_+_52719936 | 11.63 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr19_+_49496782 | 11.24 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr22_-_50219548 | 10.91 |
ENST00000404034.1
|
BRD1
|
bromodomain containing 1 |
| chr1_+_207262881 | 10.41 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_-_51425902 | 10.41 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr18_+_3449695 | 9.94 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr20_-_33872518 | 9.73 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr20_-_33872548 | 9.56 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr12_-_121019165 | 9.50 |
ENST00000341039.2
ENST00000357500.4 |
POP5
|
processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr17_-_29151794 | 9.09 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr2_+_85804614 | 9.08 |
ENST00000263864.5
ENST00000409760.1 |
VAMP8
|
vesicle-associated membrane protein 8 |
| chr1_+_181057638 | 9.06 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr11_-_107729887 | 8.78 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr19_+_49497121 | 8.67 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr12_+_69004619 | 8.57 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr16_-_30457048 | 8.53 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr2_-_61765315 | 8.35 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr19_-_13044494 | 8.10 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr3_-_33481835 | 8.02 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr9_-_132805430 | 8.01 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chr18_-_54305658 | 7.84 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr18_+_3449821 | 7.44 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_207262540 | 7.17 |
ENST00000452902.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr9_-_130635741 | 7.07 |
ENST00000223836.10
|
AK1
|
adenylate kinase 1 |
| chr1_-_225615599 | 7.06 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr1_+_207262578 | 6.97 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr22_-_37640277 | 6.61 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr13_-_46716969 | 6.48 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_74701062 | 6.27 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr17_-_30228678 | 6.20 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr1_+_207262627 | 6.17 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr17_-_79827808 | 6.11 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr1_-_70671216 | 5.97 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr12_+_72148614 | 5.86 |
ENST00000261263.3
|
RAB21
|
RAB21, member RAS oncogene family |
| chr1_+_207262170 | 5.86 |
ENST00000367078.3
|
C4BPB
|
complement component 4 binding protein, beta |
| chr2_+_219081817 | 5.66 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr4_-_100871506 | 5.55 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr6_-_32140886 | 5.51 |
ENST00000395496.1
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr5_+_43603229 | 5.34 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr16_-_3030407 | 5.21 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr9_-_123605177 | 5.20 |
ENST00000373904.5
ENST00000210313.3 |
PSMD5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr3_+_52828805 | 5.19 |
ENST00000416872.2
ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr7_-_104909435 | 5.01 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr11_-_46867780 | 4.91 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr22_-_37640456 | 4.90 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr3_-_3221358 | 4.87 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr4_-_105416039 | 4.80 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr15_-_61521495 | 4.60 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr6_+_138188551 | 4.45 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr5_-_169725231 | 4.45 |
ENST00000046794.5
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr10_+_70661014 | 4.42 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr9_-_33447584 | 4.37 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr5_-_179051579 | 4.29 |
ENST00000505811.1
ENST00000515714.1 ENST00000513225.1 ENST00000503664.1 ENST00000356731.5 ENST00000523137.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr17_+_76374714 | 4.23 |
ENST00000262764.6
ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1
|
phosphatidylglycerophosphate synthase 1 |
| chr12_+_122326662 | 4.16 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr18_+_3451584 | 4.16 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_24286287 | 4.02 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr20_+_60878005 | 4.02 |
ENST00000253003.2
|
ADRM1
|
adhesion regulating molecule 1 |
| chr1_+_28844648 | 4.01 |
ENST00000373832.1
ENST00000373831.3 |
RCC1
|
regulator of chromosome condensation 1 |
| chr2_+_198380289 | 3.95 |
ENST00000233892.4
ENST00000409916.1 |
MOB4
|
MOB family member 4, phocein |
| chr17_+_8191815 | 3.87 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr5_-_180666570 | 3.82 |
ENST00000509535.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr12_-_31479045 | 3.81 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr21_-_34185944 | 3.67 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr12_+_10658489 | 3.64 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr6_-_101329191 | 3.61 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr8_-_117768023 | 3.47 |
ENST00000518949.1
ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H
|
eukaryotic translation initiation factor 3, subunit H |
| chr11_+_74660278 | 3.47 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr7_+_76139741 | 3.44 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr3_+_111260954 | 3.40 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr3_-_52569023 | 3.34 |
ENST00000307076.4
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr10_-_73848531 | 3.30 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr2_+_191208196 | 3.12 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr13_-_41837620 | 3.09 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr11_-_111741994 | 3.08 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr6_+_31674639 | 2.98 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr1_+_109756523 | 2.97 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr10_+_51565188 | 2.93 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr4_-_46391805 | 2.90 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr16_+_30212378 | 2.88 |
ENST00000569485.1
|
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr19_+_9938562 | 2.88 |
ENST00000586895.1
ENST00000358666.3 ENST00000590068.1 ENST00000593087.1 |
UBL5
|
ubiquitin-like 5 |
| chr1_+_32479430 | 2.85 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr16_+_50776021 | 2.82 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr6_-_131949200 | 2.78 |
ENST00000539158.1
ENST00000368058.1 |
MED23
|
mediator complex subunit 23 |
| chr2_+_99953816 | 2.74 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr2_+_10560147 | 2.65 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chrX_-_153775426 | 2.63 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr19_-_50083803 | 2.63 |
ENST00000391853.3
ENST00000339093.3 |
NOSIP
|
nitric oxide synthase interacting protein |
| chr2_+_233925064 | 2.59 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr19_-_50083822 | 2.58 |
ENST00000596358.1
|
NOSIP
|
nitric oxide synthase interacting protein |
| chr15_+_68346501 | 2.57 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr6_+_150070831 | 2.53 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr6_-_24911195 | 2.52 |
ENST00000259698.4
|
FAM65B
|
family with sequence similarity 65, member B |
| chr2_+_223725652 | 2.43 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_-_145715565 | 2.38 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr8_-_141774467 | 2.33 |
ENST00000520151.1
ENST00000519024.1 ENST00000519465.1 |
PTK2
|
protein tyrosine kinase 2 |
| chr3_+_183892635 | 2.32 |
ENST00000427072.1
ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr6_+_150070857 | 2.31 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr20_-_36156293 | 2.31 |
ENST00000373537.2
ENST00000414542.2 |
BLCAP
|
bladder cancer associated protein |
| chr20_-_36156125 | 2.30 |
ENST00000397135.1
ENST00000397137.1 |
BLCAP
|
bladder cancer associated protein |
| chr10_-_16859442 | 2.30 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr18_+_3451646 | 2.28 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_203103281 | 2.26 |
ENST00000392244.3
ENST00000409181.1 ENST00000409712.1 ENST00000409498.2 ENST00000409368.1 ENST00000392245.1 ENST00000392246.2 |
SUMO1
|
small ubiquitin-like modifier 1 |
| chr1_-_33502441 | 2.16 |
ENST00000548033.1
ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2
|
adenylate kinase 2 |
| chr14_-_105635090 | 2.13 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr2_+_234668894 | 2.12 |
ENST00000305208.5
ENST00000608383.1 ENST00000360418.3 |
UGT1A8
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr14_-_94856987 | 2.12 |
ENST00000449399.3
ENST00000404814.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr12_+_62654155 | 2.09 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr12_+_95867727 | 2.04 |
ENST00000323666.5
ENST00000546753.1 |
METAP2
|
methionyl aminopeptidase 2 |
| chr17_+_38296576 | 2.02 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr1_-_35658736 | 2.00 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr19_-_12512062 | 1.96 |
ENST00000595766.1
ENST00000430385.3 |
ZNF799
|
zinc finger protein 799 |
| chr5_-_52405564 | 1.94 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr22_-_30234218 | 1.93 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr3_+_111260856 | 1.89 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr10_-_73848764 | 1.88 |
ENST00000317376.4
ENST00000412663.1 |
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr9_-_100459639 | 1.84 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr3_+_38017264 | 1.82 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_+_16484691 | 1.81 |
ENST00000344087.4
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr17_-_64225508 | 1.81 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr19_+_49375649 | 1.74 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr1_-_33502528 | 1.73 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chr14_-_94856951 | 1.70 |
ENST00000553327.1
ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr4_-_175443484 | 1.67 |
ENST00000514584.1
ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr12_-_30887948 | 1.62 |
ENST00000433722.2
|
CAPRIN2
|
caprin family member 2 |
| chrX_+_135730297 | 1.61 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr14_-_94857004 | 1.60 |
ENST00000557492.1
ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr19_-_39322299 | 1.55 |
ENST00000601094.1
ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr13_+_73302047 | 1.54 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr12_-_112847354 | 1.54 |
ENST00000550566.2
ENST00000553213.2 ENST00000424576.2 ENST00000202773.9 |
RPL6
|
ribosomal protein L6 |
| chr3_-_49823941 | 1.47 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr14_-_21994525 | 1.47 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr6_-_131949305 | 1.44 |
ENST00000368053.4
ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23
|
mediator complex subunit 23 |
| chr2_-_225811747 | 1.42 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_-_37068530 | 1.41 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr6_-_86099898 | 1.36 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr14_-_100842588 | 1.34 |
ENST00000556645.1
ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr2_-_24149977 | 1.31 |
ENST00000238789.5
|
ATAD2B
|
ATPase family, AAA domain containing 2B |
| chr16_+_27413483 | 1.31 |
ENST00000337929.3
ENST00000564089.1 |
IL21R
|
interleukin 21 receptor |
| chr15_+_49170083 | 1.27 |
ENST00000530028.2
|
EID1
|
EP300 interacting inhibitor of differentiation 1 |
| chr22_-_50946113 | 1.23 |
ENST00000216080.5
ENST00000474879.2 ENST00000380796.3 |
LMF2
|
lipase maturation factor 2 |
| chr7_+_99971068 | 1.23 |
ENST00000198536.2
ENST00000453419.1 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
| chrX_+_148622513 | 1.22 |
ENST00000393985.3
ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A
|
chromosome X open reading frame 40A |
| chr7_-_156803329 | 1.20 |
ENST00000252971.6
|
MNX1
|
motor neuron and pancreas homeobox 1 |
| chr4_+_110736659 | 1.19 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr21_+_30502806 | 1.19 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr12_-_57522813 | 1.12 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr1_-_246729544 | 1.10 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr6_-_31651817 | 1.09 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr11_-_842509 | 1.07 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr4_+_74347400 | 1.07 |
ENST00000226355.3
|
AFM
|
afamin |
| chr15_-_37391614 | 1.06 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chrX_+_135730373 | 1.06 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr1_+_154966058 | 1.06 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr1_+_2036149 | 1.04 |
ENST00000482686.1
ENST00000400920.1 ENST00000486681.1 |
PRKCZ
|
protein kinase C, zeta |
| chr16_+_31044812 | 1.03 |
ENST00000313843.3
|
STX4
|
syntaxin 4 |
| chr15_-_43785303 | 1.01 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr11_-_64052111 | 1.01 |
ENST00000394532.3
ENST00000394531.3 ENST00000309032.3 |
BAD
|
BCL2-associated agonist of cell death |
| chr9_+_34652164 | 1.01 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr16_-_57831676 | 0.99 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr12_+_104458235 | 0.96 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr15_+_49913175 | 0.94 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr3_+_63897605 | 0.94 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr16_-_20817753 | 0.93 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr16_-_31076332 | 0.92 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr3_+_45636219 | 0.91 |
ENST00000273317.4
|
LIMD1
|
LIM domains containing 1 |
| chr11_-_2924720 | 0.91 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr2_-_86790593 | 0.89 |
ENST00000263856.4
ENST00000409225.2 |
CHMP3
|
charged multivesicular body protein 3 |
| chr17_-_35969409 | 0.87 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr12_+_59989918 | 0.84 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr17_+_39411636 | 0.83 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr2_-_183106641 | 0.82 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr15_+_49913201 | 0.82 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr6_-_35656685 | 0.82 |
ENST00000539068.1
ENST00000540787.1 |
FKBP5
|
FK506 binding protein 5 |
| chr21_-_34144157 | 0.82 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr17_+_68071389 | 0.82 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr12_+_4699244 | 0.81 |
ENST00000540757.2
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr2_-_203103185 | 0.80 |
ENST00000409205.1
|
SUMO1
|
small ubiquitin-like modifier 1 |
| chr1_+_171217677 | 0.79 |
ENST00000402921.2
|
FMO1
|
flavin containing monooxygenase 1 |
| chr5_-_58882219 | 0.76 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr16_+_4896659 | 0.76 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr19_-_39322497 | 0.75 |
ENST00000221418.4
|
ECH1
|
enoyl CoA hydratase 1, peroxisomal |
| chr19_-_51537982 | 0.74 |
ENST00000525263.1
|
KLK12
|
kallikrein-related peptidase 12 |
| chr19_-_51538148 | 0.74 |
ENST00000319590.4
ENST00000250351.4 |
KLK12
|
kallikrein-related peptidase 12 |
| chr4_+_57774042 | 0.73 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr5_+_71014990 | 0.71 |
ENST00000296777.4
|
CARTPT
|
CART prepropeptide |
| chr1_+_161736072 | 0.70 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr2_+_234959323 | 0.69 |
ENST00000373368.1
ENST00000168148.3 |
SPP2
|
secreted phosphoprotein 2, 24kDa |
| chr1_-_22469459 | 0.69 |
ENST00000290167.6
|
WNT4
|
wingless-type MMTV integration site family, member 4 |
| chr7_+_76139833 | 0.69 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr10_-_11574274 | 0.65 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr14_-_106573756 | 0.65 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr7_+_99971129 | 0.63 |
ENST00000394000.2
ENST00000350573.2 |
PILRA
|
paired immunoglobin-like type 2 receptor alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 35.0 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 5.6 | 16.9 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 4.8 | 19.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 4.4 | 13.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 4.1 | 36.6 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 3.9 | 19.6 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 3.0 | 9.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 2.9 | 11.5 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 2.0 | 8.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 1.9 | 9.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.8 | 5.3 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 1.6 | 8.0 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.5 | 4.5 | GO:0034146 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 1.3 | 3.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 1.2 | 8.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.1 | 4.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 1.0 | 11.5 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 1.0 | 5.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.0 | 3.9 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.0 | 9.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.9 | 14.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.9 | 2.8 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.9 | 4.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.9 | 2.6 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.8 | 2.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.8 | 4.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.8 | 5.5 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.7 | 2.8 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.7 | 10.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.6 | 3.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.6 | 5.9 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.6 | 1.7 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.5 | 4.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 2.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.5 | 4.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.5 | 4.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 13.9 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.4 | 2.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.4 | 2.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 4.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.4 | 6.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.4 | 5.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.4 | 5.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.4 | 4.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 1.0 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 2.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.3 | 8.6 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.3 | 6.3 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.3 | 4.6 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.3 | 2.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.3 | 1.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 1.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 2.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.3 | 1.0 | GO:0052042 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.3 | 21.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 1.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 5.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.7 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.2 | 1.9 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 1.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.7 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.2 | 3.1 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 10.9 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.2 | 3.4 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 7.1 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.2 | 2.0 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.2 | 3.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 1.0 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 2.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.2 | 1.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.2 | 3.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 1.8 | GO:0051918 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 4.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.2 | 6.2 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 2.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 2.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.7 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 1.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 1.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.5 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 1.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 4.6 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 5.5 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.9 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 5.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 9.1 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 13.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.2 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.1 | 0.8 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.9 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.4 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 4.4 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 9.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 1.6 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 10.7 | GO:0002576 | platelet degranulation(GO:0002576) |
| 0.1 | 0.7 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 4.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.4 | GO:0050663 | cytokine secretion(GO:0050663) |
| 0.1 | 4.6 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.1 | 2.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.8 | GO:0018212 | peptidyl-tyrosine phosphorylation(GO:0018108) peptidyl-tyrosine modification(GO:0018212) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 1.1 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 2.9 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0035701 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 2.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 3.7 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 3.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 3.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 1.3 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 2.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 3.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.9 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 35.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 1.9 | 5.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.4 | 9.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.2 | 6.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.1 | 16.9 | GO:0045120 | pronucleus(GO:0045120) |
| 1.1 | 10.9 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.9 | 5.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.9 | 4.4 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.8 | 3.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.7 | 10.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.7 | 5.9 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.7 | 8.3 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.6 | 1.9 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.6 | 19.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.5 | 3.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.5 | 9.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.4 | 4.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.4 | 4.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 3.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.4 | 36.6 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.3 | 6.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.3 | 6.6 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.3 | 21.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.3 | 4.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.3 | 9.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 1.8 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.2 | 2.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 17.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.2 | 2.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.2 | 1.2 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 19.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.2 | 3.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 2.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 8.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 3.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 2.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 5.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 3.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.9 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 5.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 3.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.7 | GO:0005912 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.8 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 4.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.7 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 7.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 17.4 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 13.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.7 | GO:0030666 | endocytic vesicle membrane(GO:0030666) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 35.0 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 4.9 | 19.6 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 3.9 | 19.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 3.6 | 14.5 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 3.1 | 3.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 3.1 | 21.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 2.8 | 8.5 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 2.4 | 7.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 2.0 | 8.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 1.9 | 16.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.6 | 4.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 1.1 | 7.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.1 | 4.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 1.0 | 3.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.0 | 3.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.9 | 11.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.9 | 2.7 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.9 | 2.6 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.9 | 2.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.8 | 9.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.8 | 23.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.8 | 8.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.7 | 7.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 1.9 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.6 | 6.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 9.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.6 | 4.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.6 | 6.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.5 | 3.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.5 | 3.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.5 | 2.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 5.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.5 | 2.9 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.5 | 3.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.4 | 4.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.4 | 4.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 1.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 11.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 3.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 7.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 10.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 1.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 1.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 7.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.3 | 1.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.3 | 5.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.7 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 13.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 1.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 5.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.6 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.2 | 2.4 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.2 | 4.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 14.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 5.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 9.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 3.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.8 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 2.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 9.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.5 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 4.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 2.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 4.4 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 11.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 9.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 11.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 10.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 11.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.6 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 2.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 2.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 4.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 1.2 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 2.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.5 | GO:0008233 | peptidase activity(GO:0008233) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 35.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.7 | 10.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.5 | 42.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 14.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 26.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 4.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 7.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 7.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 6.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 11.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 6.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 4.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 34.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.9 | 35.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.9 | 26.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 23.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.5 | 2.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.5 | 13.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 11.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.5 | 13.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 10.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.3 | 12.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.3 | 11.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 4.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 5.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 5.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 9.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 9.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 3.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 7.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 5.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 2.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 5.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 9.4 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 3.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 10.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.8 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 4.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 4.5 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.1 | 7.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 5.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 6.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.7 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 4.0 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 4.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.2 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 3.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |