Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
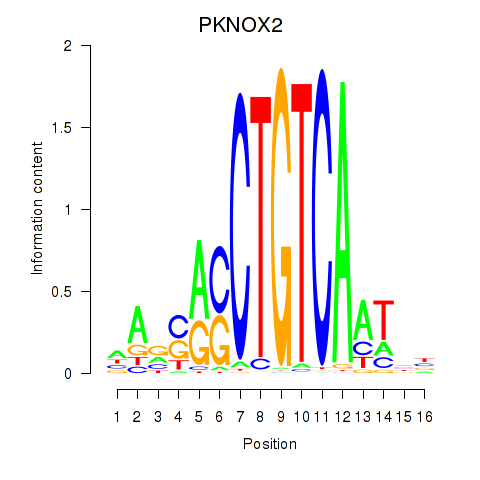
Results for PKNOX2
Z-value: 0.74
Transcription factors associated with PKNOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX2
|
ENSG00000165495.11 | PBX/knotted 1 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PKNOX2 | hg19_v2_chr11_+_125034586_125034604 | -0.41 | 3.4e-10 | Click! |
Activity profile of PKNOX2 motif
Sorted Z-values of PKNOX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_111195973 | 13.37 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr8_+_11666649 | 13.03 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_61765315 | 10.51 |
ENST00000406957.1
ENST00000401558.2 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr11_+_74660278 | 9.90 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr4_-_100871506 | 9.65 |
ENST00000296417.5
|
H2AFZ
|
H2A histone family, member Z |
| chr12_+_49621658 | 9.61 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr18_-_54318353 | 9.59 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr4_-_157892498 | 9.58 |
ENST00000502773.1
|
PDGFC
|
platelet derived growth factor C |
| chr18_-_54305658 | 9.55 |
ENST00000586262.1
ENST00000217515.6 |
TXNL1
|
thioredoxin-like 1 |
| chr7_-_148580563 | 9.42 |
ENST00000476773.1
|
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr18_+_3449695 | 9.15 |
ENST00000343820.5
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_-_64013288 | 8.60 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_+_52719936 | 8.33 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr1_+_181057638 | 8.08 |
ENST00000367577.4
|
IER5
|
immediate early response 5 |
| chr19_+_36142147 | 8.02 |
ENST00000590618.1
|
COX6B1
|
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr6_+_150070831 | 8.00 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr6_-_101329191 | 7.43 |
ENST00000324723.6
ENST00000369162.2 ENST00000522650.1 |
ASCC3
|
activating signal cointegrator 1 complex subunit 3 |
| chr17_-_29151794 | 7.27 |
ENST00000324238.6
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr3_-_33481835 | 7.05 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr6_-_32140886 | 7.04 |
ENST00000395496.1
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr16_-_30457048 | 6.96 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chrX_-_77225135 | 6.84 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr18_+_3449821 | 6.83 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr20_+_43514320 | 6.35 |
ENST00000372839.3
ENST00000428262.1 ENST00000445830.1 |
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chr6_+_150070857 | 6.31 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chrX_-_153775426 | 5.89 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr1_-_35658736 | 5.69 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr2_+_219081817 | 5.23 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr5_-_52405564 | 5.23 |
ENST00000510818.2
ENST00000396954.3 ENST00000508922.1 ENST00000361377.4 ENST00000582677.1 ENST00000584946.1 ENST00000450852.3 |
MOCS2
|
molybdenum cofactor synthesis 2 |
| chr2_+_219524473 | 5.22 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr20_-_61847586 | 4.92 |
ENST00000370339.3
|
YTHDF1
|
YTH domain family, member 1 |
| chr11_-_46867780 | 4.48 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr10_+_51565188 | 4.47 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr11_-_111741994 | 4.20 |
ENST00000398006.2
|
ALG9
|
ALG9, alpha-1,2-mannosyltransferase |
| chr4_+_140222609 | 3.78 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr15_-_37392086 | 3.51 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr17_-_27405875 | 3.39 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr6_+_29910301 | 3.17 |
ENST00000376809.5
ENST00000376802.2 |
HLA-A
|
major histocompatibility complex, class I, A |
| chr16_-_3030407 | 3.04 |
ENST00000431515.2
ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr5_+_125758865 | 2.96 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr22_-_30234218 | 2.88 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr10_+_70661014 | 2.76 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr14_-_21994525 | 2.71 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr16_+_57481382 | 2.71 |
ENST00000564655.1
ENST00000567072.1 ENST00000567933.1 ENST00000563166.1 |
COQ9
|
coenzyme Q9 |
| chr3_-_127541194 | 2.63 |
ENST00000453507.2
|
MGLL
|
monoglyceride lipase |
| chr1_+_161736072 | 2.62 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr1_+_74701062 | 2.62 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_+_109756523 | 2.60 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr6_+_26124373 | 2.58 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr5_+_125758813 | 2.57 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr9_-_132805430 | 2.51 |
ENST00000446176.2
ENST00000355681.3 ENST00000420781.1 |
FNBP1
|
formin binding protein 1 |
| chr22_-_31885514 | 2.46 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr16_+_57481349 | 2.37 |
ENST00000262507.6
ENST00000565964.1 |
COQ9
|
coenzyme Q9 |
| chr4_+_169418195 | 2.32 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr2_-_202316260 | 2.20 |
ENST00000332624.3
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr8_+_57124245 | 2.05 |
ENST00000521831.1
ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr11_-_842509 | 2.05 |
ENST00000322028.4
|
POLR2L
|
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr16_-_57481278 | 1.98 |
ENST00000567751.1
ENST00000568940.1 ENST00000563341.1 ENST00000565961.1 ENST00000569370.1 ENST00000567518.1 ENST00000565786.1 ENST00000394391.4 |
CIAPIN1
|
cytokine induced apoptosis inhibitor 1 |
| chr6_-_116575226 | 1.98 |
ENST00000420283.1
|
TSPYL4
|
TSPY-like 4 |
| chr6_+_17600576 | 1.96 |
ENST00000259963.3
|
FAM8A1
|
family with sequence similarity 8, member A1 |
| chr4_-_46391805 | 1.95 |
ENST00000540012.1
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr7_-_104909435 | 1.94 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr11_+_842808 | 1.73 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr5_-_94620239 | 1.70 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_31674639 | 1.64 |
ENST00000556581.1
ENST00000375832.4 ENST00000503322.1 |
LY6G6F
MEGT1
|
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr12_-_121734489 | 1.56 |
ENST00000412367.2
ENST00000402834.4 ENST00000404169.3 |
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr3_-_127541679 | 1.54 |
ENST00000265052.5
|
MGLL
|
monoglyceride lipase |
| chr5_-_81046922 | 1.40 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr5_-_81046841 | 1.38 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr11_+_842928 | 1.37 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr6_-_87804815 | 1.36 |
ENST00000369582.2
|
CGA
|
glycoprotein hormones, alpha polypeptide |
| chr3_-_3221358 | 1.27 |
ENST00000424814.1
ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN
|
cereblon |
| chr8_+_133787586 | 1.22 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr2_+_191208196 | 1.18 |
ENST00000392329.2
ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr19_+_10828724 | 1.16 |
ENST00000585892.1
ENST00000314646.5 ENST00000359692.6 |
DNM2
|
dynamin 2 |
| chr12_+_62654119 | 1.12 |
ENST00000353364.3
ENST00000549523.1 ENST00000280377.5 |
USP15
|
ubiquitin specific peptidase 15 |
| chr3_+_52828805 | 1.00 |
ENST00000416872.2
ENST00000449956.2 |
ITIH3
|
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr21_-_34185944 | 0.98 |
ENST00000479548.1
|
C21orf62
|
chromosome 21 open reading frame 62 |
| chr11_+_12132117 | 0.87 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr13_+_33160553 | 0.74 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr19_+_41281060 | 0.69 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr3_+_32147997 | 0.65 |
ENST00000282541.5
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr7_-_137028498 | 0.59 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr12_+_62654155 | 0.58 |
ENST00000312635.6
ENST00000393654.3 ENST00000549237.1 |
USP15
|
ubiquitin specific peptidase 15 |
| chr19_+_41281282 | 0.46 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr1_-_179112173 | 0.41 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr12_-_91398796 | 0.41 |
ENST00000261172.3
ENST00000551767.1 |
EPYC
|
epiphycan |
| chr5_+_118407053 | 0.37 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr18_+_54318616 | 0.32 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr19_+_7733929 | 0.29 |
ENST00000221515.2
|
RETN
|
resistin |
| chr19_+_41281416 | 0.28 |
ENST00000597140.1
|
MIA
|
melanoma inhibitory activity |
| chr6_+_138188551 | 0.27 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_+_10828795 | 0.23 |
ENST00000389253.4
ENST00000355667.6 ENST00000408974.4 |
DNM2
|
dynamin 2 |
| chr2_+_64681103 | 0.20 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr5_-_81046904 | 0.20 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr6_+_108881012 | 0.19 |
ENST00000343882.6
|
FOXO3
|
forkhead box O3 |
| chr17_+_68071389 | 0.05 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr19_-_6424783 | 0.04 |
ENST00000398148.3
|
KHSRP
|
KH-type splicing regulatory protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 3.1 | 9.4 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 2.6 | 13.0 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 2.4 | 9.6 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 2.0 | 5.9 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 1.7 | 6.8 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.6 | 14.3 | GO:0030091 | protein repair(GO:0030091) |
| 1.5 | 10.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 1.5 | 10.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.9 | 5.7 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.7 | 2.2 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.7 | 5.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.7 | 5.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.6 | 3.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.6 | 4.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 7.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.5 | 2.6 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.5 | 7.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.5 | 1.4 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.5 | 9.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.4 | 4.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 1.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 | 1.6 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.4 | 1.9 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 8.0 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.3 | 3.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 7.7 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 | 0.9 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 0.3 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.3 | 5.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.2 | 5.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 4.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 19.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.2 | 0.6 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 9.7 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 4.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.4 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 4.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 6.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 2.6 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.1 | 1.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 2.0 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.3 | GO:2000870 | positive regulation of female gonad development(GO:2000196) regulation of progesterone secretion(GO:2000870) |
| 0.1 | 1.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 7.3 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 8.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 2.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 3.5 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 2.0 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 7.0 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 3.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 2.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 3.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 10.0 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.5 | GO:0019827 | stem cell population maintenance(GO:0019827) maintenance of cell number(GO:0098727) |
| 0.0 | 1.2 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 1.4 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.2 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.7 | 5.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 1.6 | 9.7 | GO:0001740 | Barr body(GO:0001740) |
| 1.4 | 9.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.9 | 10.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.7 | 5.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.6 | 9.4 | GO:0045120 | pronucleus(GO:0045120) |
| 0.5 | 3.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 5.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 4.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 3.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 6.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 8.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 19.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.1 | 4.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 12.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 2.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 2.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.0 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 2.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 6.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 26.6 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 2.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 3.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 5.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.4 | GO:0005796 | Golgi lumen(GO:0005796) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 3.3 | 13.0 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 2.7 | 19.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 2.3 | 7.0 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 2.0 | 5.9 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 1.7 | 5.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 1.7 | 6.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.4 | 4.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 1.0 | 9.4 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.0 | 10.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.9 | 7.4 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.9 | 6.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.9 | 2.6 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.6 | 16.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.4 | 9.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 4.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 4.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 7.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.3 | 3.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.3 | 1.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 8.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 13.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 2.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.2 | 9.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 9.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.2 | 3.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 6.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 4.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 1.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 2.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 2.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 12.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 5.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.7 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 4.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 6.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 20.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 13.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 9.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 3.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 3.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 8.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.5 | 13.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.5 | 6.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.5 | 9.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 13.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 13.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 9.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 16.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 3.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 7.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 12.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 2.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 4.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 4.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 8.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 5.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 2.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 5.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 2.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 4.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 3.5 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |