Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
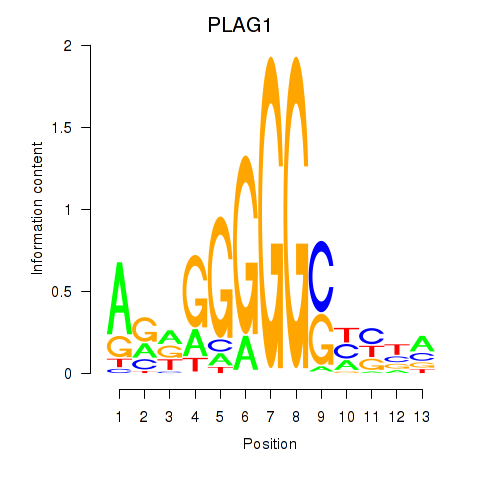
Results for PLAG1
Z-value: 1.05
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.3 | PLAG1 zinc finger |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PLAG1 | hg19_v2_chr8_-_57123815_57123867 | 0.30 | 4.9e-06 | Click! |
Activity profile of PLAG1 motif
Sorted Z-values of PLAG1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_139874683 | 29.76 |
ENST00000444903.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr12_+_79258444 | 19.04 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr10_-_21786179 | 17.42 |
ENST00000377113.5
|
CASC10
|
cancer susceptibility candidate 10 |
| chr12_+_79258547 | 17.06 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr8_+_21912328 | 15.13 |
ENST00000432128.1
ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN
|
dematin actin binding protein |
| chr12_+_57522258 | 14.02 |
ENST00000553277.1
ENST00000243077.3 |
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr7_+_72742162 | 13.50 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr7_-_158380371 | 12.16 |
ENST00000389418.4
ENST00000389416.4 |
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr7_+_72742178 | 11.60 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr15_+_74833518 | 11.60 |
ENST00000346246.5
|
ARID3B
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr11_-_2160180 | 11.54 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr11_-_111170526 | 11.33 |
ENST00000355430.4
|
COLCA1
|
colorectal cancer associated 1 |
| chr3_+_32726774 | 11.33 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chrX_+_110339439 | 11.29 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chrX_+_51927919 | 10.97 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr11_-_6440283 | 10.70 |
ENST00000299402.6
ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr6_-_33285505 | 10.32 |
ENST00000431845.2
|
ZBTB22
|
zinc finger and BTB domain containing 22 |
| chr5_+_161274940 | 10.28 |
ENST00000393943.4
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr19_+_40697514 | 10.20 |
ENST00000253055.3
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr7_-_158380465 | 10.17 |
ENST00000389413.3
ENST00000409483.1 |
PTPRN2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr18_-_51750948 | 9.69 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr6_-_150039249 | 9.29 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr15_+_29131103 | 9.21 |
ENST00000558402.1
ENST00000558330.1 |
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr14_-_93799360 | 9.14 |
ENST00000334746.5
ENST00000554565.1 ENST00000298896.3 |
BTBD7
|
BTB (POZ) domain containing 7 |
| chrX_-_45060135 | 8.58 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr12_-_122018859 | 8.54 |
ENST00000536437.1
ENST00000377071.4 ENST00000538046.2 |
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr1_+_163038565 | 8.22 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr20_-_18038521 | 7.95 |
ENST00000278780.6
|
OVOL2
|
ovo-like zinc finger 2 |
| chr10_-_75571566 | 7.94 |
ENST00000299641.4
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr12_-_112819896 | 7.64 |
ENST00000377560.5
ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr1_-_31845914 | 7.51 |
ENST00000373713.2
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_-_5890145 | 7.20 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr12_-_52715179 | 7.18 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr14_+_57857262 | 7.17 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr11_-_45687128 | 7.14 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr11_-_73309228 | 7.12 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr5_+_161275320 | 7.02 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr1_+_22962948 | 6.84 |
ENST00000374642.3
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr12_+_107168342 | 6.77 |
ENST00000392837.4
|
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr10_-_75571341 | 6.72 |
ENST00000309979.6
|
NDST2
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr22_+_29469100 | 6.58 |
ENST00000327813.5
ENST00000407188.1 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr16_+_29674277 | 6.54 |
ENST00000395389.2
|
SPN
|
sialophorin |
| chrX_-_137793826 | 6.48 |
ENST00000315930.6
|
FGF13
|
fibroblast growth factor 13 |
| chr14_-_75079026 | 6.32 |
ENST00000261978.4
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr2_-_152955213 | 6.21 |
ENST00000427385.1
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr17_+_73106035 | 6.19 |
ENST00000581078.1
ENST00000582136.1 ENST00000245543.1 |
ARMC7
|
armadillo repeat containing 7 |
| chr11_-_57282349 | 6.19 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr20_+_18269121 | 6.17 |
ENST00000377671.3
ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133
|
zinc finger protein 133 |
| chr11_+_46299199 | 5.99 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr6_-_31940065 | 5.84 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr16_-_65155833 | 5.67 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chrY_+_22737604 | 5.54 |
ENST00000361365.2
|
EIF1AY
|
eukaryotic translation initiation factor 1A, Y-linked |
| chr10_-_99447024 | 5.41 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr17_-_4607335 | 5.37 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr9_-_123476612 | 5.32 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr9_-_123476719 | 5.30 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr16_+_31128978 | 5.20 |
ENST00000448516.2
ENST00000219797.4 |
KAT8
|
K(lysine) acetyltransferase 8 |
| chr2_-_30144432 | 5.17 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr9_-_135819987 | 4.91 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr2_-_21266935 | 4.77 |
ENST00000233242.1
|
APOB
|
apolipoprotein B |
| chr16_-_29910853 | 4.74 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr1_-_110613276 | 4.71 |
ENST00000369792.4
|
ALX3
|
ALX homeobox 3 |
| chr19_-_2702681 | 4.65 |
ENST00000382159.3
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chrX_-_78622805 | 4.63 |
ENST00000373298.2
|
ITM2A
|
integral membrane protein 2A |
| chr14_+_79746249 | 4.59 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr16_+_2198604 | 4.55 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr3_-_52001448 | 4.51 |
ENST00000461554.1
ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4
|
poly(rC) binding protein 4 |
| chr22_+_29469012 | 4.50 |
ENST00000400335.4
ENST00000400338.2 |
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr10_+_111767720 | 4.46 |
ENST00000356080.4
ENST00000277900.8 |
ADD3
|
adducin 3 (gamma) |
| chr1_-_20306909 | 4.46 |
ENST00000375111.3
ENST00000400520.3 |
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr2_-_86850949 | 4.28 |
ENST00000237455.4
|
RNF103
|
ring finger protein 103 |
| chr4_+_55095264 | 4.28 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr14_+_79745746 | 4.26 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr2_-_152955537 | 4.25 |
ENST00000201943.5
ENST00000539935.1 |
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_-_59189545 | 4.24 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr9_-_140115775 | 4.14 |
ENST00000391553.1
ENST00000392827.1 |
RNF208
|
ring finger protein 208 |
| chrX_-_71526999 | 4.10 |
ENST00000453707.2
ENST00000373619.3 |
CITED1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr14_+_79745682 | 4.01 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr12_-_12419703 | 3.96 |
ENST00000543091.1
ENST00000261349.4 |
LRP6
|
low density lipoprotein receptor-related protein 6 |
| chr17_+_48046538 | 3.88 |
ENST00000240306.3
|
DLX4
|
distal-less homeobox 4 |
| chr2_-_178937478 | 3.85 |
ENST00000286063.6
|
PDE11A
|
phosphodiesterase 11A |
| chr11_-_64511789 | 3.84 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_-_1420006 | 3.76 |
ENST00000320345.6
ENST00000406424.4 |
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr15_+_92397051 | 3.60 |
ENST00000424469.2
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1 |
| chr3_+_14989076 | 3.54 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_+_16080659 | 3.49 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr12_+_121163538 | 3.47 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr20_+_35169885 | 3.41 |
ENST00000279022.2
ENST00000346786.2 |
MYL9
|
myosin, light chain 9, regulatory |
| chr16_-_30134524 | 3.37 |
ENST00000395202.1
ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3
|
mitogen-activated protein kinase 3 |
| chr3_-_73673991 | 3.33 |
ENST00000308537.4
ENST00000263666.4 |
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr17_+_7284365 | 3.24 |
ENST00000311668.2
|
TNK1
|
tyrosine kinase, non-receptor, 1 |
| chr12_-_108733078 | 3.22 |
ENST00000552995.1
ENST00000312143.7 ENST00000397688.2 ENST00000550402.1 |
CMKLR1
|
chemokine-like receptor 1 |
| chr3_-_197024394 | 3.12 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr20_-_1974692 | 3.09 |
ENST00000217305.2
ENST00000539905.1 |
PDYN
|
prodynorphin |
| chr6_+_106534192 | 3.07 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr2_-_74618964 | 3.06 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr3_+_19988885 | 2.93 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr16_+_29984962 | 2.92 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr22_+_19744226 | 2.68 |
ENST00000332710.4
ENST00000329705.7 ENST00000359500.3 |
TBX1
|
T-box 1 |
| chr1_-_204654481 | 2.61 |
ENST00000367176.3
|
LRRN2
|
leucine rich repeat neuronal 2 |
| chr11_+_66610883 | 2.47 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr3_+_124303539 | 2.44 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr13_+_53602894 | 2.34 |
ENST00000219022.2
|
OLFM4
|
olfactomedin 4 |
| chr2_+_157292933 | 2.19 |
ENST00000540309.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr3_-_15374033 | 2.18 |
ENST00000253688.5
ENST00000383791.3 |
SH3BP5
|
SH3-domain binding protein 5 (BTK-associated) |
| chr6_+_7727030 | 2.14 |
ENST00000283147.6
|
BMP6
|
bone morphogenetic protein 6 |
| chr9_-_34662651 | 2.13 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr10_+_50817141 | 2.10 |
ENST00000339797.1
|
CHAT
|
choline O-acetyltransferase |
| chr1_+_2407754 | 1.99 |
ENST00000419816.2
ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2
|
phospholipase C, eta 2 |
| chr16_-_29910365 | 1.92 |
ENST00000346932.5
ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr9_-_136004782 | 1.90 |
ENST00000393157.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr2_-_233352531 | 1.74 |
ENST00000304546.1
|
ECEL1
|
endothelin converting enzyme-like 1 |
| chr13_-_49107303 | 1.61 |
ENST00000344532.3
|
RCBTB2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr11_+_1940925 | 1.59 |
ENST00000453458.1
ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3
|
troponin T type 3 (skeletal, fast) |
| chr17_-_50237374 | 1.52 |
ENST00000442502.2
|
CA10
|
carbonic anhydrase X |
| chr2_-_128399706 | 1.47 |
ENST00000426981.1
|
LIMS2
|
LIM and senescent cell antigen-like domains 2 |
| chr19_+_45973120 | 1.46 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr10_+_102891048 | 1.32 |
ENST00000467928.2
|
TLX1
|
T-cell leukemia homeobox 1 |
| chr1_-_201438282 | 1.28 |
ENST00000367311.3
ENST00000367309.1 |
PHLDA3
|
pleckstrin homology-like domain, family A, member 3 |
| chr1_-_115632035 | 1.27 |
ENST00000433172.1
ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2
|
tetraspanin 2 |
| chr1_-_92949505 | 1.25 |
ENST00000370332.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr7_-_22233442 | 1.25 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr14_+_32546485 | 1.25 |
ENST00000345122.3
ENST00000432921.1 ENST00000433497.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr12_-_58131931 | 1.14 |
ENST00000547588.1
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr6_+_31939608 | 1.07 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr20_+_35201857 | 1.00 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr6_-_71012773 | 0.85 |
ENST00000370496.3
ENST00000357250.6 |
COL9A1
|
collagen, type IX, alpha 1 |
| chr2_+_220306745 | 0.76 |
ENST00000431523.1
ENST00000396698.1 ENST00000396695.2 |
SPEG
|
SPEG complex locus |
| chr2_-_74619152 | 0.76 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr11_+_63997750 | 0.76 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr4_-_38666430 | 0.68 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr1_+_202431859 | 0.57 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_+_57998595 | 0.45 |
ENST00000337737.3
ENST00000548198.1 ENST00000551632.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr14_-_36989427 | 0.44 |
ENST00000354822.5
|
NKX2-1
|
NK2 homeobox 1 |
| chr19_+_38924316 | 0.43 |
ENST00000355481.4
ENST00000360985.3 ENST00000359596.3 |
RYR1
|
ryanodine receptor 1 (skeletal) |
| chr7_+_73703728 | 0.41 |
ENST00000361545.5
ENST00000223398.6 |
CLIP2
|
CAP-GLY domain containing linker protein 2 |
| chr4_-_155533787 | 0.32 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr1_-_27701307 | 0.32 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr19_-_18653781 | 0.24 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr12_+_57998400 | 0.21 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr17_-_3819751 | 0.16 |
ENST00000225538.3
|
P2RX1
|
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr16_-_18812746 | 0.11 |
ENST00000546206.2
ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1
RP11-1035H13.3
|
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr14_-_106518922 | 0.10 |
ENST00000390598.2
|
IGHV3-7
|
immunoglobulin heavy variable 3-7 |
| chr3_+_19988566 | 0.07 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr14_-_25519317 | 0.06 |
ENST00000323944.5
|
STXBP6
|
syntaxin binding protein 6 (amisyn) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PLAG1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.0 | 36.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 4.7 | 14.0 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 3.6 | 10.7 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 3.1 | 25.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.8 | 8.5 | GO:0021678 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 2.5 | 15.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 2.5 | 7.5 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 2.4 | 7.2 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 2.0 | 7.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 2.0 | 4.0 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 1.9 | 9.3 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 1.7 | 29.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.6 | 6.5 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 1.5 | 10.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 1.4 | 4.3 | GO:0072275 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.3 | 6.5 | GO:1990834 | response to odorant(GO:1990834) |
| 1.3 | 5.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 1.3 | 11.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.2 | 14.7 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 1.2 | 7.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 1.2 | 4.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 1.0 | 4.9 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.9 | 3.8 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.9 | 2.7 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.9 | 17.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 12.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.7 | 4.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.7 | 4.8 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.7 | 3.4 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.7 | 6.0 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.6 | 9.7 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.6 | 3.1 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.6 | 11.3 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.6 | 5.7 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.5 | 2.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.5 | 3.0 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.5 | 2.4 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.5 | 3.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.5 | 4.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.4 | 3.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.4 | 2.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.4 | 1.3 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.4 | 21.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 5.8 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.4 | 7.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.3 | 5.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 2.3 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.3 | 4.5 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.2 | 3.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 3.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 8.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.3 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 13.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 3.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.2 | 3.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.2 | 1.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 15.8 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 6.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 6.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 6.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 7.1 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 4.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 4.7 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.1 | 2.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 2.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.2 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 5.4 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 2.9 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.4 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 4.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 9.4 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 10.2 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.1 | 1.5 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.1 | 0.9 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.1 | 7.2 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 1.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 11.1 | GO:0060173 | appendage development(GO:0048736) limb development(GO:0060173) |
| 0.1 | 1.7 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 3.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 3.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 2.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0042692 | muscle cell differentiation(GO:0042692) |
| 0.0 | 3.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 4.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 7.6 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 1.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 2.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 2.0 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) positive regulation of amino acid transport(GO:0051957) |
| 0.0 | 3.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.1 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 36.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 1.8 | 7.2 | GO:0031417 | NatC complex(GO:0031417) |
| 1.4 | 6.8 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 1.3 | 4.0 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.2 | 4.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.2 | 10.7 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.9 | 15.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.8 | 25.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.7 | 11.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.6 | 4.8 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.5 | 6.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.5 | 17.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.5 | 3.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.4 | 3.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.4 | 2.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 6.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 27.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 15.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 22.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 3.1 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 3.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 5.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 0.9 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 3.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 10.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 9.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 11.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 7.5 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.1 | 6.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 28.7 | GO:0098793 | presynapse(GO:0098793) |
| 0.1 | 4.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 8.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 4.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 3.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 9.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 7.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 3.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 7.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 2.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 2.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 5.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 13.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.6 | GO:0031672 | A band(GO:0031672) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 29.8 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 7.2 | 36.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 3.7 | 14.7 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 3.5 | 14.0 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 2.6 | 10.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 2.5 | 7.5 | GO:0070538 | oleic acid binding(GO:0070538) |
| 2.2 | 17.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 1.9 | 5.8 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 1.4 | 4.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.2 | 4.8 | GO:0035473 | lipase binding(GO:0035473) |
| 1.1 | 8.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 1.0 | 7.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.0 | 9.7 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.9 | 25.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 22.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.8 | 12.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.7 | 5.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.6 | 3.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.6 | 3.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 10.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.6 | 7.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.5 | 2.2 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.5 | 2.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.5 | 2.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.5 | 14.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.5 | 10.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.5 | 15.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.5 | 6.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 4.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 3.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 5.2 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.4 | 7.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.4 | 1.6 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.3 | 4.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.3 | 11.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 15.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 4.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 6.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 3.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 11.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 4.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 1.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 4.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.2 | 3.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 3.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 6.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 13.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 2.9 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 2.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 6.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 6.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 8.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 1.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.9 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 2.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 3.6 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 3.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 3.2 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.1 | 2.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 11.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 3.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 5.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 6.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 4.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 4.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 8.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.3 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 39.5 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.5 | 15.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.4 | 13.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 14.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 4.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 11.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.2 | 14.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 5.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 4.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 5.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 3.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 3.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 4.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 5.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 7.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 2.2 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 11.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 36.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.6 | 6.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.6 | 17.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 9.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.4 | 11.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.4 | 14.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 2.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 9.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 4.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.3 | 3.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.3 | 25.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.3 | 7.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 6.2 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 4.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 7.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.2 | 7.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 3.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 4.8 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 5.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 11.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 3.6 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 6.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 3.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.9 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 3.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 2.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 3.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 2.2 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |