Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for PRDM1
Z-value: 0.07
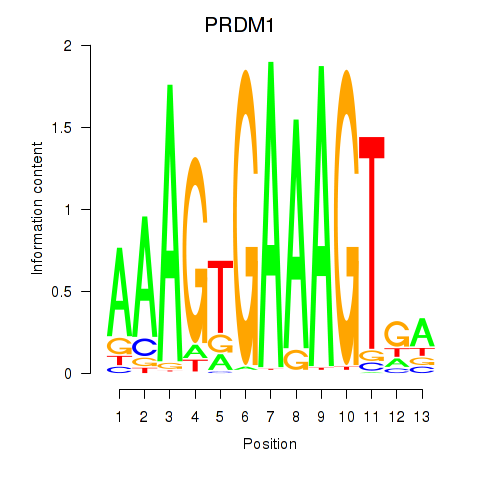
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PR/SET domain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106534192_106534224 | 0.16 | 2.0e-02 | Click! |
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_32821924 | 10.92 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_-_111746966 | 8.48 |
ENST00000369752.5
|
DENND2D
|
DENN/MADD domain containing 2D |
| chr17_-_26694979 | 8.13 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr17_-_26695013 | 8.12 |
ENST00000555059.2
|
CTB-96E2.2
|
Homeobox protein SEBOX |
| chr17_-_79818354 | 7.54 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr6_+_32811885 | 6.96 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr16_-_67970990 | 6.84 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr14_+_24605361 | 6.72 |
ENST00000206451.6
ENST00000559123.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr6_-_33282163 | 6.53 |
ENST00000434618.2
ENST00000456592.2 |
TAPBP
|
TAP binding protein (tapasin) |
| chr14_+_24605389 | 5.92 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_-_231084617 | 5.89 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr10_-_44880491 | 5.71 |
ENST00000374426.2
ENST00000395795.4 ENST00000395794.2 ENST00000374429.2 ENST00000395793.3 ENST00000343575.6 |
CXCL12
|
chemokine (C-X-C motif) ligand 12 |
| chr12_+_25205446 | 5.43 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr2_-_231084820 | 5.36 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr3_-_49851313 | 5.16 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr16_+_50776021 | 5.07 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr6_-_32811771 | 4.94 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr16_+_50775971 | 4.83 |
ENST00000311559.9
ENST00000564326.1 ENST00000566206.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr16_+_50775948 | 4.83 |
ENST00000569681.1
ENST00000569418.1 ENST00000540145.1 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr12_-_57505121 | 4.59 |
ENST00000538913.2
ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr22_-_31688431 | 4.54 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_-_21490417 | 4.42 |
ENST00000556366.1
|
NDRG2
|
NDRG family member 2 |
| chr22_-_36556821 | 4.39 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chr2_-_231084659 | 4.39 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr10_-_82049424 | 4.32 |
ENST00000372213.3
|
MAT1A
|
methionine adenosyltransferase I, alpha |
| chr5_+_49962495 | 4.25 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr11_+_46402482 | 4.24 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_118827333 | 4.18 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr6_-_41909191 | 4.17 |
ENST00000512426.1
ENST00000372987.4 |
CCND3
|
cyclin D3 |
| chr16_-_88851618 | 4.08 |
ENST00000301015.9
|
PIEZO1
|
piezo-type mechanosensitive ion channel component 1 |
| chr7_-_86849025 | 4.07 |
ENST00000257637.3
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr13_+_31309645 | 3.96 |
ENST00000380490.3
|
ALOX5AP
|
arachidonate 5-lipoxygenase-activating protein |
| chr5_+_96211643 | 3.96 |
ENST00000437043.3
ENST00000510373.1 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr4_-_100242549 | 3.87 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr5_-_39274617 | 3.72 |
ENST00000510188.1
|
FYB
|
FYN binding protein |
| chr5_+_96212185 | 3.68 |
ENST00000379904.4
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr9_-_34710066 | 3.61 |
ENST00000378792.1
ENST00000259607.2 |
CCL21
|
chemokine (C-C motif) ligand 21 |
| chr3_+_8543533 | 3.53 |
ENST00000454244.1
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr2_+_231191875 | 3.44 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr14_-_21490653 | 3.39 |
ENST00000449431.2
|
NDRG2
|
NDRG family member 2 |
| chr22_-_31688381 | 3.16 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr19_+_24009879 | 3.07 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr3_+_8543561 | 2.96 |
ENST00000397386.3
|
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr2_+_231280908 | 2.93 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr11_+_128563652 | 2.77 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_+_97506033 | 2.66 |
ENST00000518385.1
|
SDC2
|
syndecan 2 |
| chr12_+_6561190 | 2.52 |
ENST00000544021.1
ENST00000266556.7 |
TAPBPL
|
TAP binding protein-like |
| chr12_-_106641728 | 2.49 |
ENST00000378026.4
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr9_+_137533615 | 2.38 |
ENST00000371817.3
|
COL5A1
|
collagen, type V, alpha 1 |
| chr5_-_39203093 | 2.35 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr22_+_39436862 | 2.34 |
ENST00000381565.2
ENST00000452957.2 |
APOBEC3F
APOBEC3G
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| chr11_+_128563948 | 2.24 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr14_-_24615805 | 2.24 |
ENST00000560410.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr11_+_128634589 | 2.23 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_+_96145974 | 2.15 |
ENST00000315367.3
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr19_+_49977818 | 2.12 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr6_-_112575912 | 2.11 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr14_-_21490590 | 2.10 |
ENST00000557633.1
|
NDRG2
|
NDRG family member 2 |
| chr17_+_41158742 | 2.08 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr3_-_114343039 | 2.08 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_+_46402583 | 2.06 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr5_-_96143602 | 2.01 |
ENST00000443439.2
ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr1_+_212782012 | 1.98 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr12_-_120763739 | 1.94 |
ENST00000549767.1
|
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr5_-_96143796 | 1.89 |
ENST00000296754.3
|
ERAP1
|
endoplasmic reticulum aminopeptidase 1 |
| chr17_+_7211656 | 1.88 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr6_-_29527702 | 1.88 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr1_-_221915418 | 1.86 |
ENST00000323825.3
ENST00000366899.3 |
DUSP10
|
dual specificity phosphatase 10 |
| chr14_-_21490958 | 1.82 |
ENST00000554104.1
|
NDRG2
|
NDRG family member 2 |
| chr6_-_137113604 | 1.78 |
ENST00000359015.4
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr3_+_8543393 | 1.67 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr10_-_50747064 | 1.64 |
ENST00000355832.5
ENST00000603152.1 ENST00000447839.2 |
ERCC6
PGBD3
ERCC6-PGBD3
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 piggyBac transposable element derived 3 ERCC6-PGBD3 readthrough |
| chr20_+_30946106 | 1.63 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr1_+_181003067 | 1.59 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr11_+_64008525 | 1.59 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr1_+_2985760 | 1.57 |
ENST00000378391.2
ENST00000514189.1 ENST00000270722.5 |
PRDM16
|
PR domain containing 16 |
| chr4_+_152020789 | 1.53 |
ENST00000512690.1
ENST00000508783.1 ENST00000512797.1 ENST00000507327.1 ENST00000515792.1 ENST00000506126.1 |
RPS3A
|
ribosomal protein S3A |
| chr13_+_102104980 | 1.53 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr7_+_134464376 | 1.50 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr2_-_175260368 | 1.47 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr4_+_152020715 | 1.47 |
ENST00000274065.4
|
RPS3A
|
ribosomal protein S3A |
| chr19_+_17186577 | 1.42 |
ENST00000595618.1
ENST00000594824.1 |
MYO9B
|
myosin IXB |
| chr4_+_152020736 | 1.40 |
ENST00000509736.1
ENST00000505243.1 ENST00000514682.1 ENST00000322686.6 ENST00000503002.1 |
RPS3A
|
ribosomal protein S3A |
| chr19_+_13106383 | 1.36 |
ENST00000397661.2
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chrX_-_154563889 | 1.36 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr20_+_388791 | 1.34 |
ENST00000441733.1
ENST00000353660.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_-_93447333 | 1.33 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr7_-_41742697 | 1.33 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr4_+_142558078 | 1.33 |
ENST00000529613.1
|
IL15
|
interleukin 15 |
| chr11_+_46402297 | 1.29 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr2_+_231280954 | 1.29 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr6_-_33281979 | 1.24 |
ENST00000426633.2
ENST00000467025.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr2_+_145780767 | 1.23 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr11_+_46402744 | 1.21 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_+_167525277 | 1.20 |
ENST00000400926.2
|
CCR6
|
chemokine (C-C motif) receptor 6 |
| chr6_-_112575687 | 1.18 |
ENST00000521398.1
ENST00000424408.2 ENST00000243219.3 |
LAMA4
|
laminin, alpha 4 |
| chr1_-_8483723 | 1.17 |
ENST00000476556.1
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr8_-_56986768 | 1.16 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr4_+_142557771 | 1.14 |
ENST00000514653.1
|
IL15
|
interleukin 15 |
| chr6_-_112575838 | 1.14 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr2_+_234545092 | 1.11 |
ENST00000344644.5
|
UGT1A10
|
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr4_+_142557717 | 1.11 |
ENST00000320650.4
ENST00000296545.7 |
IL15
|
interleukin 15 |
| chr4_-_100140331 | 1.10 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr6_+_26156551 | 1.10 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr6_-_112575758 | 1.07 |
ENST00000431543.2
ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4
|
laminin, alpha 4 |
| chr6_-_167276033 | 1.06 |
ENST00000503859.1
ENST00000506565.1 |
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr6_+_118869452 | 1.03 |
ENST00000357525.5
|
PLN
|
phospholamban |
| chr12_-_86650077 | 1.02 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr2_+_234590556 | 1.02 |
ENST00000373426.3
|
UGT1A7
|
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr6_-_132910877 | 1.01 |
ENST00000258034.2
|
TAAR5
|
trace amine associated receptor 5 |
| chr3_-_149375783 | 1.01 |
ENST00000467467.1
ENST00000460517.1 ENST00000360632.3 |
WWTR1
|
WW domain containing transcription regulator 1 |
| chr17_+_6659153 | 1.01 |
ENST00000441631.1
ENST00000438512.1 ENST00000346752.4 ENST00000361842.3 |
XAF1
|
XIAP associated factor 1 |
| chr1_+_2985726 | 1.01 |
ENST00000511072.1
ENST00000378398.3 ENST00000441472.2 ENST00000442529.2 |
PRDM16
|
PR domain containing 16 |
| chr11_+_64008443 | 1.00 |
ENST00000309366.4
|
FKBP2
|
FK506 binding protein 2, 13kDa |
| chr18_-_47017956 | 0.99 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr16_-_3930724 | 0.98 |
ENST00000262367.5
|
CREBBP
|
CREB binding protein |
| chr11_+_64323428 | 0.98 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr8_-_29120580 | 0.98 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr5_-_64064508 | 0.98 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr6_+_152011628 | 0.95 |
ENST00000404742.1
ENST00000440973.1 |
ESR1
|
estrogen receptor 1 |
| chr11_+_64323156 | 0.95 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr3_+_69915385 | 0.95 |
ENST00000314589.5
|
MITF
|
microphthalmia-associated transcription factor |
| chr14_-_23479331 | 0.92 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr12_+_9144626 | 0.92 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chrX_+_123095546 | 0.91 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr22_+_41697520 | 0.90 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr6_-_32806506 | 0.85 |
ENST00000374897.2
ENST00000452392.2 |
TAP2
TAP2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr1_-_44497118 | 0.81 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_+_207262881 | 0.80 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr10_-_35104185 | 0.80 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr12_-_86650045 | 0.80 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_-_44497024 | 0.80 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr7_-_122526499 | 0.80 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr12_+_51318513 | 0.78 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr1_-_153919128 | 0.76 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr15_-_37393406 | 0.76 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr2_+_175260451 | 0.74 |
ENST00000458563.1
ENST00000409673.3 ENST00000272732.6 ENST00000435964.1 |
SCRN3
|
secernin 3 |
| chrX_-_106362013 | 0.73 |
ENST00000372487.1
ENST00000372479.3 ENST00000203616.8 |
RBM41
|
RNA binding motif protein 41 |
| chr5_-_149380698 | 0.73 |
ENST00000296736.3
|
TIGD6
|
tigger transposable element derived 6 |
| chr15_+_85144217 | 0.69 |
ENST00000540936.1
ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr20_+_388935 | 0.69 |
ENST00000382181.2
ENST00000400247.3 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_+_112227311 | 0.68 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr20_+_388679 | 0.66 |
ENST00000356286.5
ENST00000475269.1 |
RBCK1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_-_111558135 | 0.64 |
ENST00000394598.2
ENST00000394595.3 |
PITX2
|
paired-like homeodomain 2 |
| chr3_+_107241783 | 0.63 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr14_-_24615523 | 0.63 |
ENST00000559056.1
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr21_+_43933946 | 0.63 |
ENST00000352133.2
|
SLC37A1
|
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr1_+_199996702 | 0.62 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr18_+_18943554 | 0.60 |
ENST00000580732.2
|
GREB1L
|
growth regulation by estrogen in breast cancer-like |
| chr21_+_22370608 | 0.60 |
ENST00000400546.1
|
NCAM2
|
neural cell adhesion molecule 2 |
| chr17_+_46908350 | 0.57 |
ENST00000258947.3
ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2
|
calcium binding and coiled-coil domain 2 |
| chr8_+_77593448 | 0.57 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr5_-_94417339 | 0.56 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_69240302 | 0.56 |
ENST00000303714.4
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr11_-_85338311 | 0.56 |
ENST00000376104.2
|
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr17_+_74372662 | 0.54 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr8_+_134203273 | 0.54 |
ENST00000250160.6
|
WISP1
|
WNT1 inducible signaling pathway protein 1 |
| chr2_+_135676381 | 0.53 |
ENST00000537343.1
ENST00000295238.6 ENST00000264157.5 |
CCNT2
|
cyclin T2 |
| chr2_+_69240415 | 0.53 |
ENST00000409829.3
|
ANTXR1
|
anthrax toxin receptor 1 |
| chr5_-_59481406 | 0.49 |
ENST00000546160.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr1_+_199996733 | 0.48 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr3_+_185046676 | 0.48 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr15_-_37392086 | 0.47 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr8_+_77593474 | 0.45 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr11_+_64323098 | 0.43 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr2_-_64246206 | 0.42 |
ENST00000409558.4
ENST00000272322.4 |
VPS54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr13_-_95364389 | 0.38 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr12_+_94542459 | 0.36 |
ENST00000258526.4
|
PLXNC1
|
plexin C1 |
| chr6_+_146348782 | 0.34 |
ENST00000361719.2
ENST00000392299.2 |
GRM1
|
glutamate receptor, metabotropic 1 |
| chr3_-_46000064 | 0.30 |
ENST00000433878.1
|
FYCO1
|
FYVE and coiled-coil domain containing 1 |
| chr3_-_168864427 | 0.30 |
ENST00000468789.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr20_+_6748311 | 0.30 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr2_+_234526272 | 0.28 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr15_+_71185148 | 0.22 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr5_+_140430979 | 0.22 |
ENST00000306549.3
|
PCDHB1
|
protocadherin beta 1 |
| chr16_+_10972818 | 0.20 |
ENST00000576601.1
|
CIITA
|
class II, major histocompatibility complex, transactivator |
| chr1_-_244006528 | 0.14 |
ENST00000336199.5
ENST00000263826.5 |
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr7_+_134832808 | 0.12 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr6_+_146348810 | 0.10 |
ENST00000492807.2
|
GRM1
|
glutamate receptor, metabotropic 1 |
| chr19_-_6670128 | 0.07 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr6_-_121655552 | 0.07 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr22_-_30642782 | 0.07 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr6_-_10412600 | 0.06 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr6_-_167275991 | 0.06 |
ENST00000510118.1
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr2_+_145780739 | 0.05 |
ENST00000597173.1
ENST00000602108.1 ENST00000420472.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr10_+_22610124 | 0.04 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr7_+_134464414 | 0.04 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr15_+_71184931 | 0.01 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr19_+_34287751 | 0.01 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr1_+_110453608 | 0.00 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 2.6 | 7.7 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 2.5 | 2.5 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 1.9 | 9.3 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.5 | 19.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 1.5 | 8.8 | GO:0030421 | defecation(GO:0030421) |
| 1.2 | 3.6 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 1.1 | 4.6 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 1.1 | 4.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.1 | 4.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 1.0 | 4.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.9 | 7.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.9 | 8.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 11.7 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.7 | 2.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.7 | 2.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.6 | 5.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.6 | 2.4 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.5 | 1.6 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.5 | 1.6 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.5 | 5.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.5 | 1.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.5 | 1.9 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.4 | 1.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.4 | 4.0 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.4 | 2.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.4 | 1.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.4 | 41.7 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.4 | 2.7 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 1.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.3 | 1.0 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.3 | 8.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 2.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 7.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 1.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.3 | 2.3 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.3 | 1.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 2.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 3.7 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.2 | 1.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 2.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 0.6 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.5 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.2 | 0.9 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.3 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.9 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 4.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.1 | 1.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.5 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.1 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 1.4 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.1 | 4.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.1 | 1.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 2.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 5.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.8 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 5.4 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 3.1 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 1.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 4.2 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0045345 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.6 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.0 | 2.8 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 5.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 29.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 2.6 | 15.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 2.5 | 7.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 2.0 | 8.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.4 | 8.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.8 | 2.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 2.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.7 | 2.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.6 | 4.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 2.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 1.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.4 | 1.3 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.3 | 14.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 1.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 5.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 5.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 2.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.4 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.1 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 2.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 1.0 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 4.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 5.4 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 5.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 11.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 4.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.0 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 4.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 7.2 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 8.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 7.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 6.2 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 4.8 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 15.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 2.1 | 8.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.5 | 7.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 1.5 | 14.7 | GO:1990380 | Lys63-specific deubiquitinase activity(GO:0061578) Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.3 | 4.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.0 | 3.9 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.9 | 26.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.9 | 5.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.8 | 5.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.7 | 4.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 4.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.5 | 8.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.5 | 2.3 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 1.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 1.6 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.4 | 1.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.4 | 7.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 5.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 1.8 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.0 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 2.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 7.7 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.2 | 0.6 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 1.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 2.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.2 | 9.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 1.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.7 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.9 | GO:0048273 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 4.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.6 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 1.0 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 1.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 3.6 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 2.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.7 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.5 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 8.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 2.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 5.7 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 1.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 2.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 5.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 4.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 9.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 13.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 2.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 3.0 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.3 | 8.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 14.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 5.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 4.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 4.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 11.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 7.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 3.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 6.1 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 4.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.1 | 2.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 2.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 8.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.6 | 41.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 4.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.4 | 19.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.3 | 7.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 2.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.2 | 6.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 10.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.0 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 4.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 4.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 5.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 8.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.0 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 4.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.0 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |