|
chr16_+_447226
Show fit
|
12.63 |
ENST00000433358.1
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4
|
|
chrX_-_151903101
Show fit
|
11.75 |
ENST00000393900.3
|
MAGEA12
|
melanoma antigen family A, 12
|
|
chrX_-_151903184
Show fit
|
11.75 |
ENST00000357916.4
ENST00000393869.3
|
MAGEA12
|
melanoma antigen family A, 12
|
|
chr16_+_447209
Show fit
|
11.55 |
ENST00000382940.4
ENST00000219479.2
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4
|
|
chrX_+_151903207
Show fit
|
10.55 |
ENST00000370287.3
|
CSAG1
|
chondrosarcoma associated gene 1
|
|
chrX_+_151867214
Show fit
|
9.03 |
ENST00000329342.5
ENST00000412733.1
ENST00000457643.1
|
MAGEA6
|
melanoma antigen family A, 6
|
|
chrX_+_151903253
Show fit
|
9.00 |
ENST00000452779.2
ENST00000370291.2
|
CSAG1
|
chondrosarcoma associated gene 1
|
|
chrX_-_151938171
Show fit
|
7.33 |
ENST00000393902.3
ENST00000417212.1
ENST00000370278.3
|
MAGEA3
|
melanoma antigen family A, 3
|
|
chrX_-_151922340
Show fit
|
6.12 |
ENST00000370284.1
ENST00000543232.1
ENST00000393876.1
ENST00000393872.3
|
MAGEA2
|
melanoma antigen family A, 2
|
|
chr1_+_46668994
Show fit
|
5.81 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1
|
|
chrX_+_151883090
Show fit
|
5.79 |
ENST00000370293.2
ENST00000423993.1
ENST00000447530.1
ENST00000458057.1
ENST00000331220.2
ENST00000422085.1
ENST00000453150.1
ENST00000409560.1
|
MAGEA2B
|
melanoma antigen family A, 2B
|
|
chrX_+_151081351
Show fit
|
4.78 |
ENST00000276344.2
|
MAGEA4
|
melanoma antigen family A, 4
|
|
chr6_+_151646800
Show fit
|
4.66 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr7_-_19157248
Show fit
|
4.64 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1
|
|
chr8_+_144816303
Show fit
|
4.41 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head)
|
|
chr19_-_6720686
Show fit
|
4.36 |
ENST00000245907.6
|
C3
|
complement component 3
|
|
chr7_+_75932863
Show fit
|
4.26 |
ENST00000429938.1
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr12_+_54447637
Show fit
|
4.26 |
ENST00000609810.1
ENST00000430889.2
|
HOXC4
HOXC4
|
homeobox C4
Homeobox protein Hox-C4
|
|
chrX_+_153813407
Show fit
|
4.09 |
ENST00000443287.2
ENST00000333128.3
|
CTAG1A
|
cancer/testis antigen 1A
|
|
chr10_+_17271266
Show fit
|
3.74 |
ENST00000224237.5
|
VIM
|
vimentin
|
|
chr11_+_71791693
Show fit
|
3.53 |
ENST00000289488.2
ENST00000447974.1
|
LRTOMT
|
leucine rich transmembrane and O-methyltransferase domain containing
|
|
chrX_-_153847522
Show fit
|
3.36 |
ENST00000328435.2
ENST00000359887.4
|
CTAG1B
|
cancer/testis antigen 1B
|
|
chrX_-_153881842
Show fit
|
3.36 |
ENST00000369585.3
ENST00000247306.4
|
CTAG2
|
cancer/testis antigen 2
|
|
chr11_+_69455855
Show fit
|
3.29 |
ENST00000227507.2
ENST00000536559.1
|
CCND1
|
cyclin D1
|
|
chrX_-_151307020
Show fit
|
3.22 |
ENST00000370323.4
ENST00000244096.3
ENST00000427322.2
|
MAGEA10
|
melanoma antigen family A, 10
|
|
chr17_-_39677971
Show fit
|
3.10 |
ENST00000393976.2
|
KRT15
|
keratin 15
|
|
chr12_+_96588279
Show fit
|
3.00 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr11_+_86511569
Show fit
|
2.96 |
ENST00000441050.1
|
PRSS23
|
protease, serine, 23
|
|
chr12_-_53074182
Show fit
|
2.89 |
ENST00000252244.3
|
KRT1
|
keratin 1
|
|
chr1_-_204380919
Show fit
|
2.82 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr4_+_74735102
Show fit
|
2.79 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha)
|
|
chr3_+_154797877
Show fit
|
2.74 |
ENST00000462745.1
ENST00000493237.1
|
MME
|
membrane metallo-endopeptidase
|
|
chr11_+_46402744
Show fit
|
2.72 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chrX_+_102631844
Show fit
|
2.68 |
ENST00000372634.1
ENST00000299872.7
|
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr12_-_56101647
Show fit
|
2.54 |
ENST00000347027.6
ENST00000257879.6
ENST00000257880.7
ENST00000394230.2
ENST00000394229.2
|
ITGA7
|
integrin, alpha 7
|
|
chr4_+_123747979
Show fit
|
2.49 |
ENST00000608478.1
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chrX_-_148669116
Show fit
|
2.48 |
ENST00000243314.5
|
MAGEA9B
|
melanoma antigen family A, 9B
|
|
chr9_-_21994597
Show fit
|
2.40 |
ENST00000579755.1
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A
|
|
chr3_+_154797428
Show fit
|
2.38 |
ENST00000460393.1
|
MME
|
membrane metallo-endopeptidase
|
|
chr2_+_46926326
Show fit
|
2.36 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chrX_-_100604184
Show fit
|
2.36 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast)
|
|
chrX_+_148863584
Show fit
|
2.33 |
ENST00000439010.2
ENST00000298974.5
ENST00000522429.1
ENST00000519822.1
|
MAGEA9
|
melanoma antigen family A, 9
|
|
chr9_-_13279563
Show fit
|
2.25 |
ENST00000541718.1
|
MPDZ
|
multiple PDZ domain protein
|
|
chr9_-_21994344
Show fit
|
2.25 |
ENST00000530628.2
ENST00000361570.3
|
CDKN2A
|
cyclin-dependent kinase inhibitor 2A
|
|
chr17_-_66453562
Show fit
|
2.25 |
ENST00000262139.5
ENST00000546360.1
|
WIPI1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chrX_+_17755563
Show fit
|
2.23 |
ENST00000380045.3
ENST00000380041.3
ENST00000380043.3
ENST00000398080.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila)
|
|
chr2_+_48757278
Show fit
|
2.19 |
ENST00000404752.1
ENST00000406226.1
|
STON1
|
stonin 1
|
|
chr9_-_13279406
Show fit
|
2.19 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein
|
|
chr8_+_28748099
Show fit
|
2.18 |
ENST00000519047.1
|
HMBOX1
|
homeobox containing 1
|
|
chr19_-_42759300
Show fit
|
2.06 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor
|
|
chr19_+_55477711
Show fit
|
2.04 |
ENST00000448584.2
ENST00000537859.1
ENST00000585500.1
ENST00000427260.2
ENST00000538819.1
ENST00000263437.6
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chr19_+_55476620
Show fit
|
2.03 |
ENST00000543010.1
ENST00000391721.4
ENST00000339757.7
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chr17_-_72864739
Show fit
|
2.03 |
ENST00000579893.1
ENST00000544854.1
|
FDXR
|
ferredoxin reductase
|
|
chr19_+_35609380
Show fit
|
2.00 |
ENST00000604621.1
|
FXYD3
|
FXYD domain containing ion transport regulator 3
|
|
chr13_-_37494365
Show fit
|
1.96 |
ENST00000350148.5
|
SMAD9
|
SMAD family member 9
|
|
chr4_+_123747834
Show fit
|
1.95 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr13_-_37494391
Show fit
|
1.94 |
ENST00000379826.4
|
SMAD9
|
SMAD family member 9
|
|
chr12_-_123849374
Show fit
|
1.91 |
ENST00000602398.1
ENST00000602750.1
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr1_-_153538292
Show fit
|
1.86 |
ENST00000497140.1
ENST00000368708.3
|
S100A2
|
S100 calcium binding protein A2
|
|
chr16_-_3285144
Show fit
|
1.85 |
ENST00000431561.3
ENST00000396870.4
|
ZNF200
|
zinc finger protein 200
|
|
chr19_-_3029011
Show fit
|
1.85 |
ENST00000590536.1
ENST00000587137.1
ENST00000455444.2
ENST00000262953.6
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr12_+_96588143
Show fit
|
1.85 |
ENST00000228741.3
ENST00000547249.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2)
|
|
chr22_+_32750872
Show fit
|
1.83 |
ENST00000397468.1
|
RFPL3
|
ret finger protein-like 3
|
|
chr1_+_18958008
Show fit
|
1.78 |
ENST00000420770.2
ENST00000400661.3
|
PAX7
|
paired box 7
|
|
chr11_+_46403303
Show fit
|
1.76 |
ENST00000407067.1
ENST00000395565.1
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr19_-_3025614
Show fit
|
1.76 |
ENST00000447365.2
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila)
|
|
chr12_+_54402790
Show fit
|
1.75 |
ENST00000040584.4
|
HOXC8
|
homeobox C8
|
|
chr11_+_46403194
Show fit
|
1.71 |
ENST00000395569.4
ENST00000395566.4
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr3_-_55521323
Show fit
|
1.68 |
ENST00000264634.4
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr21_+_44073916
Show fit
|
1.64 |
ENST00000349112.3
ENST00000398224.3
|
PDE9A
|
phosphodiesterase 9A
|
|
chr9_-_13279589
Show fit
|
1.63 |
ENST00000319217.7
|
MPDZ
|
multiple PDZ domain protein
|
|
chr9_-_14314066
Show fit
|
1.61 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B
|
|
chr9_-_14313893
Show fit
|
1.60 |
ENST00000380921.3
ENST00000380959.3
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_7541808
Show fit
|
1.55 |
ENST00000379802.3
|
DSP
|
desmoplakin
|
|
chr1_+_244998602
Show fit
|
1.55 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor
|
|
chr9_+_132427883
Show fit
|
1.52 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2
|
|
chr21_+_44073860
Show fit
|
1.51 |
ENST00000335512.4
ENST00000539837.1
ENST00000291539.6
ENST00000380328.2
ENST00000398232.3
ENST00000398234.3
ENST00000398236.3
ENST00000328862.6
ENST00000335440.6
ENST00000398225.3
ENST00000398229.3
ENST00000398227.3
|
PDE9A
|
phosphodiesterase 9A
|
|
chr22_-_46450024
Show fit
|
1.48 |
ENST00000396008.2
ENST00000333761.1
|
C22orf26
|
chromosome 22 open reading frame 26
|
|
chr6_+_7541845
Show fit
|
1.48 |
ENST00000418664.2
|
DSP
|
desmoplakin
|
|
chr16_-_4588469
Show fit
|
1.46 |
ENST00000588381.1
ENST00000563332.2
|
CDIP1
|
cell death-inducing p53 target 1
|
|
chr16_-_31439735
Show fit
|
1.46 |
ENST00000287490.4
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr16_+_28889703
Show fit
|
1.45 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr18_+_55102917
Show fit
|
1.44 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2
|
|
chr16_+_28834303
Show fit
|
1.43 |
ENST00000340394.8
ENST00000325215.6
ENST00000395547.2
ENST00000336783.4
ENST00000382686.4
ENST00000564304.1
|
ATXN2L
|
ataxin 2-like
|
|
chr10_+_115438920
Show fit
|
1.42 |
ENST00000429617.1
ENST00000369331.4
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase
|
|
chr1_+_154975110
Show fit
|
1.41 |
ENST00000535420.1
ENST00000368426.3
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr1_-_113249948
Show fit
|
1.39 |
ENST00000339083.7
ENST00000369642.3
|
RHOC
|
ras homolog family member C
|
|
chr13_+_60971427
Show fit
|
1.39 |
ENST00000535286.1
ENST00000377881.2
|
TDRD3
|
tudor domain containing 3
|
|
chr19_-_51456198
Show fit
|
1.39 |
ENST00000594846.1
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr16_+_28889801
Show fit
|
1.38 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr13_-_74708372
Show fit
|
1.38 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12
|
|
chr19_+_3366547
Show fit
|
1.38 |
ENST00000341919.3
ENST00000590282.1
ENST00000443272.2
|
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor)
|
|
chr4_+_147560042
Show fit
|
1.34 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2
|
|
chr7_+_114562172
Show fit
|
1.34 |
ENST00000393486.1
ENST00000257724.3
|
MDFIC
|
MyoD family inhibitor domain containing
|
|
chr19_-_51456321
Show fit
|
1.33 |
ENST00000391809.2
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr12_+_16064258
Show fit
|
1.33 |
ENST00000524480.1
ENST00000531803.1
ENST00000532964.1
|
DERA
|
deoxyribose-phosphate aldolase (putative)
|
|
chr21_-_47648665
Show fit
|
1.31 |
ENST00000450351.1
ENST00000522411.1
ENST00000356396.4
ENST00000397728.3
ENST00000457828.2
|
LSS
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase)
|
|
chr1_-_113249734
Show fit
|
1.29 |
ENST00000484054.3
ENST00000369636.2
ENST00000369637.1
ENST00000285735.2
ENST00000369638.2
|
RHOC
|
ras homolog family member C
|
|
chr1_-_113249678
Show fit
|
1.29 |
ENST00000369633.2
ENST00000425265.2
ENST00000369632.2
ENST00000436685.2
|
RHOC
|
ras homolog family member C
|
|
chrX_-_131352152
Show fit
|
1.28 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family
|
|
chr1_-_17307173
Show fit
|
1.27 |
ENST00000438542.1
ENST00000375535.3
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr19_-_19051927
Show fit
|
1.26 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila)
|
|
chr16_+_28834531
Show fit
|
1.25 |
ENST00000570200.1
|
ATXN2L
|
ataxin 2-like
|
|
chr7_-_24797032
Show fit
|
1.25 |
ENST00000409970.1
ENST00000409775.3
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chrX_+_119495934
Show fit
|
1.23 |
ENST00000218008.3
ENST00000361319.3
ENST00000539306.1
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr2_+_105471969
Show fit
|
1.21 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3
|
|
chr1_-_113392399
Show fit
|
1.21 |
ENST00000449572.2
ENST00000433505.1
|
RP11-426L16.8
|
RP11-426L16.8
|
|
chr11_+_111896320
Show fit
|
1.21 |
ENST00000531306.1
ENST00000537636.1
|
DLAT
|
dihydrolipoamide S-acetyltransferase
|
|
chr15_+_63481668
Show fit
|
1.18 |
ENST00000321437.4
ENST00000559006.1
ENST00000448330.2
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr15_-_35088340
Show fit
|
1.18 |
ENST00000290378.4
|
ACTC1
|
actin, alpha, cardiac muscle 1
|
|
chr19_+_48867652
Show fit
|
1.18 |
ENST00000344846.2
|
SYNGR4
|
synaptogyrin 4
|
|
chr12_-_131323754
Show fit
|
1.17 |
ENST00000261653.6
|
STX2
|
syntaxin 2
|
|
chr5_+_150591678
Show fit
|
1.17 |
ENST00000523466.1
|
GM2A
|
GM2 ganglioside activator
|
|
chr10_+_13628933
Show fit
|
1.16 |
ENST00000417658.1
ENST00000320054.4
|
PRPF18
|
pre-mRNA processing factor 18
|
|
chr19_+_51358166
Show fit
|
1.14 |
ENST00000601503.1
ENST00000326003.2
ENST00000597286.1
ENST00000597483.1
|
KLK3
|
kallikrein-related peptidase 3
|
|
chr15_-_43513187
Show fit
|
1.14 |
ENST00000540029.1
ENST00000441366.2
|
EPB42
|
erythrocyte membrane protein band 4.2
|
|
chr14_+_74353320
Show fit
|
1.13 |
ENST00000540593.1
ENST00000555730.1
|
ZNF410
|
zinc finger protein 410
|
|
chr19_+_51358191
Show fit
|
1.12 |
ENST00000593997.1
ENST00000595952.1
ENST00000360617.3
ENST00000598145.1
|
KLK3
|
kallikrein-related peptidase 3
|
|
chr19_-_15560730
Show fit
|
1.11 |
ENST00000389282.4
ENST00000263381.7
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr6_+_7107830
Show fit
|
1.11 |
ENST00000379933.3
|
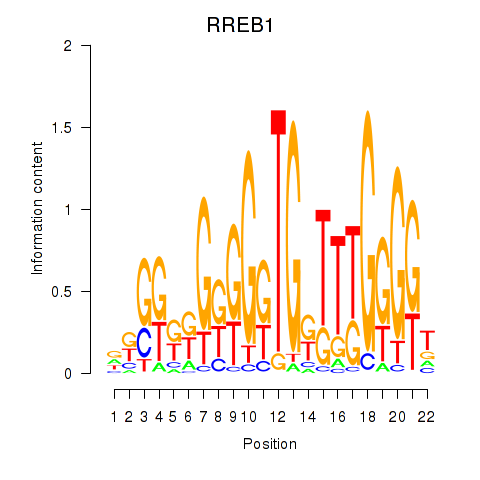
RREB1
|
ras responsive element binding protein 1
|
|
chr19_-_51456344
Show fit
|
1.11 |
ENST00000336334.3
ENST00000593428.1
|
KLK5
|
kallikrein-related peptidase 5
|
|
chr17_+_77751931
Show fit
|
1.10 |
ENST00000310942.4
ENST00000269399.5
|
CBX2
|
chromobox homolog 2
|
|
chr13_-_76056250
Show fit
|
1.08 |
ENST00000377636.3
ENST00000431480.2
ENST00000377625.2
ENST00000425511.1
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr16_+_69599861
Show fit
|
1.07 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr19_+_36157715
Show fit
|
1.06 |
ENST00000379013.2
ENST00000222275.2
|
UPK1A
|
uroplakin 1A
|
|
chr16_+_1728305
Show fit
|
1.06 |
ENST00000569765.1
|
HN1L
|
hematological and neurological expressed 1-like
|
|
chr3_+_186435137
Show fit
|
1.06 |
ENST00000447445.1
|
KNG1
|
kininogen 1
|
|
chr17_-_58156273
Show fit
|
1.06 |
ENST00000593228.1
ENST00000585976.1
ENST00000593097.1
ENST00000184956.6
|
HEATR6
|
HEAT repeat containing 6
|
|
chr2_+_192542850
Show fit
|
1.05 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1
|
|
chr8_+_32406137
Show fit
|
1.03 |
ENST00000521670.1
|
NRG1
|
neuregulin 1
|
|
chr6_+_168227611
Show fit
|
1.02 |
ENST00000344191.4
ENST00000351017.4
ENST00000392108.3
ENST00000366806.2
ENST00000392112.1
ENST00000400824.4
ENST00000447894.2
ENST00000400822.3
|
MLLT4
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4
|
|
chr12_-_56120865
Show fit
|
1.02 |
ENST00000548898.1
ENST00000552067.1
|
CD63
|
CD63 molecule
|
|
chr18_+_9334786
Show fit
|
1.00 |
ENST00000581641.1
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1
|
|
chr11_-_74109422
Show fit
|
0.99 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr17_+_79373540
Show fit
|
0.97 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1
|
|
chr12_-_44152551
Show fit
|
0.97 |
ENST00000416848.2
ENST00000550784.1
ENST00000547156.1
ENST00000549868.1
ENST00000553166.1
ENST00000551923.1
ENST00000431332.3
ENST00000344862.5
|
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like
|
|
chr12_-_131323719
Show fit
|
0.97 |
ENST00000392373.2
|
STX2
|
syntaxin 2
|
|
chr4_-_96470350
Show fit
|
0.96 |
ENST00000504962.1
ENST00000453304.1
ENST00000506749.1
|
UNC5C
|
unc-5 homolog C (C. elegans)
|
|
chr20_+_48599506
Show fit
|
0.95 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1
|
|
chr19_-_51222707
Show fit
|
0.95 |
ENST00000391814.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr10_-_75415825
Show fit
|
0.95 |
ENST00000394810.2
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr12_-_56120838
Show fit
|
0.94 |
ENST00000548160.1
|
CD63
|
CD63 molecule
|
|
chr19_+_7828035
Show fit
|
0.94 |
ENST00000327325.5
ENST00000394122.2
ENST00000248228.4
ENST00000334806.5
ENST00000359059.5
ENST00000357361.2
ENST00000596363.1
ENST00000595751.1
ENST00000596707.1
ENST00000597522.1
ENST00000595496.1
|
CLEC4M
|
C-type lectin domain family 4, member M
|
|
chr9_+_131174024
Show fit
|
0.92 |
ENST00000420034.1
ENST00000372842.1
|
CERCAM
|
cerebral endothelial cell adhesion molecule
|
|
chr16_+_68771128
Show fit
|
0.92 |
ENST00000261769.5
ENST00000422392.2
|
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial)
|
|
chr9_-_14722715
Show fit
|
0.91 |
ENST00000380911.3
|
CER1
|
cerberus 1, DAN family BMP antagonist
|
|
chr2_+_85843252
Show fit
|
0.90 |
ENST00000409025.1
ENST00000409470.1
ENST00000323701.6
ENST00000409766.3
|
USP39
|
ubiquitin specific peptidase 39
|
|
chr2_-_54087066
Show fit
|
0.89 |
ENST00000394705.2
ENST00000352846.3
ENST00000406625.2
|
GPR75
GPR75-ASB3
ASB3
|
G protein-coupled receptor 75
GPR75-ASB3 readthrough
Ankyrin repeat and SOCS box protein 3
|
|
chr1_+_154975258
Show fit
|
0.89 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr17_+_41177220
Show fit
|
0.89 |
ENST00000587250.2
ENST00000544533.1
|
RND2
|
Rho family GTPase 2
|
|
chr1_-_228604328
Show fit
|
0.89 |
ENST00000355586.4
ENST00000366698.2
ENST00000520264.1
ENST00000479800.1
ENST00000295033.3
|
TRIM17
|
tripartite motif containing 17
|
|
chr4_+_190992087
Show fit
|
0.88 |
ENST00000553598.1
|
DUX4L7
|
double homeobox 4 like 7
|
|
chr2_+_223289208
Show fit
|
0.88 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2
|
|
chr1_+_18957500
Show fit
|
0.86 |
ENST00000375375.3
|
PAX7
|
paired box 7
|
|
chr2_-_220108309
Show fit
|
0.86 |
ENST00000409640.1
|
GLB1L
|
galactosidase, beta 1-like
|
|
chr18_+_9334755
Show fit
|
0.86 |
ENST00000262120.5
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1
|
|
chr11_-_89224488
Show fit
|
0.85 |
ENST00000534731.1
ENST00000527626.1
|
NOX4
|
NADPH oxidase 4
|
|
chr19_-_51192661
Show fit
|
0.85 |
ENST00000391813.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1
|
|
chr1_+_157963063
Show fit
|
0.83 |
ENST00000360089.4
ENST00000368173.3
ENST00000392272.2
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chrX_+_149737046
Show fit
|
0.82 |
ENST00000370396.2
ENST00000542741.1
ENST00000543350.1
ENST00000424519.1
ENST00000413012.2
|
MTM1
|
myotubularin 1
|
|
chr8_+_28748765
Show fit
|
0.82 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1
|
|
chr1_-_201346761
Show fit
|
0.80 |
ENST00000455702.1
ENST00000422165.1
ENST00000367318.5
ENST00000367320.2
ENST00000438742.1
ENST00000412633.1
ENST00000458432.2
ENST00000421663.2
ENST00000367322.1
ENST00000509001.1
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr1_-_156470556
Show fit
|
0.80 |
ENST00000489057.1
ENST00000348159.4
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr8_+_73449625
Show fit
|
0.80 |
ENST00000523207.1
|
KCNB2
|
potassium voltage-gated channel, Shab-related subfamily, member 2
|
|
chr5_+_151151504
Show fit
|
0.79 |
ENST00000356245.3
ENST00000507878.2
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr18_+_33877654
Show fit
|
0.79 |
ENST00000257209.4
ENST00000445677.1
ENST00000590592.1
ENST00000359247.4
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr17_-_46703826
Show fit
|
0.76 |
ENST00000550387.1
ENST00000311177.5
|
HOXB9
|
homeobox B9
|
|
chr19_+_44085189
Show fit
|
0.75 |
ENST00000562365.2
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing
|
|
chr20_+_57875658
Show fit
|
0.75 |
ENST00000371025.3
|
EDN3
|
endothelin 3
|
|
chr7_+_91570165
Show fit
|
0.75 |
ENST00000356239.3
ENST00000359028.2
ENST00000358100.2
|
AKAP9
|
A kinase (PRKA) anchor protein 9
|
|
chr16_+_67198683
Show fit
|
0.75 |
ENST00000517685.1
ENST00000521374.1
ENST00000584272.1
|
HSF4
|
heat shock transcription factor 4
|
|
chr7_-_154794763
Show fit
|
0.74 |
ENST00000404141.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1
|
|
chr3_+_192958914
Show fit
|
0.73 |
ENST00000264735.2
ENST00000602513.1
|
HRASLS
|
HRAS-like suppressor
|
|
chr7_+_39125365
Show fit
|
0.73 |
ENST00000559001.1
ENST00000464276.2
|
POU6F2
|
POU class 6 homeobox 2
|
|
chr17_-_62658186
Show fit
|
0.73 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2
|
|
chr3_+_8775466
Show fit
|
0.73 |
ENST00000343849.2
ENST00000397368.2
|
CAV3
|
caveolin 3
|
|
chr18_+_77160282
Show fit
|
0.72 |
ENST00000318065.5
ENST00000545796.1
ENST00000592223.1
ENST00000329101.4
ENST00000586434.1
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr17_+_35849937
Show fit
|
0.72 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14
|
|
chr12_-_6716569
Show fit
|
0.72 |
ENST00000544040.1
ENST00000545942.1
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr8_+_32406179
Show fit
|
0.71 |
ENST00000405005.3
|
NRG1
|
neuregulin 1
|
|
chr5_+_151151471
Show fit
|
0.71 |
ENST00000394123.3
ENST00000543466.1
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr20_-_25062767
Show fit
|
0.70 |
ENST00000429762.3
ENST00000444511.2
ENST00000376707.3
|
VSX1
|
visual system homeobox 1
|
|
chr17_-_1733114
Show fit
|
0.68 |
ENST00000305513.7
|
SMYD4
|
SET and MYND domain containing 4
|
|
chr7_-_38398721
Show fit
|
0.68 |
ENST00000390346.2
|
TRGV3
|
T cell receptor gamma variable 3
|
|
chr4_+_190992387
Show fit
|
0.68 |
ENST00000554906.2
ENST00000553820.2
|
DUX4L7
|
double homeobox 4 like 7
|
|
chr1_-_161193349
Show fit
|
0.68 |
ENST00000469730.2
ENST00000463273.1
ENST00000464492.1
ENST00000367990.3
ENST00000470459.2
ENST00000468465.1
ENST00000463812.1
|
APOA2
|
apolipoprotein A-II
|
|
chr19_+_44084696
Show fit
|
0.67 |
ENST00000562255.1
ENST00000569031.2
|
PINLYP
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing
|
|
chr16_+_21169976
Show fit
|
0.67 |
ENST00000572258.1
ENST00000261388.3
ENST00000451578.2
ENST00000572599.1
ENST00000577162.1
|
TMEM159
|
transmembrane protein 159
|
|
chr3_+_141205852
Show fit
|
0.67 |
ENST00000286364.3
ENST00000452898.1
|
RASA2
|
RAS p21 protein activator 2
|
|
chr2_+_28618532
Show fit
|
0.67 |
ENST00000545753.1
|
FOSL2
|
FOS-like antigen 2
|
|
chrX_+_53078273
Show fit
|
0.66 |
ENST00000332582.4
|
GPR173
|
G protein-coupled receptor 173
|
|
chr4_+_164265035
Show fit
|
0.66 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5
|
|
chr10_-_35379241
Show fit
|
0.66 |
ENST00000374748.1
ENST00000374749.3
|
CUL2
|
cullin 2
|
|
chr7_-_14028488
Show fit
|
0.66 |
ENST00000405358.4
|
ETV1
|
ets variant 1
|
|
chr11_-_96239990
Show fit
|
0.65 |
ENST00000511243.2
|
JRKL-AS1
|
JRKL antisense RNA 1
|
|
chr6_-_35480705
Show fit
|
0.64 |
ENST00000229771.6
|
TULP1
|
tubby like protein 1
|
|
chr5_-_35195338
Show fit
|
0.64 |
ENST00000509839.1
|
PRLR
|
prolactin receptor
|
|
chr1_-_156470515
Show fit
|
0.64 |
ENST00000340875.5
ENST00000368240.2
ENST00000353795.3
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr10_-_121356518
Show fit
|
0.63 |
ENST00000369092.4
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr7_-_154794621
Show fit
|
0.63 |
ENST00000419436.1
ENST00000397192.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1
|