Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
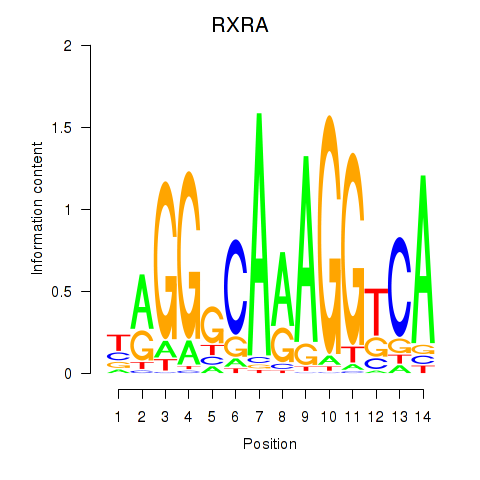
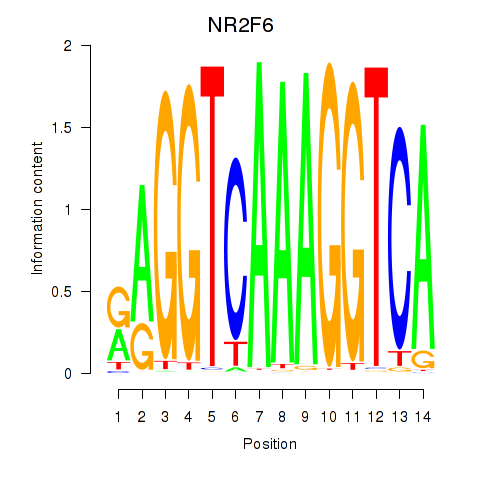
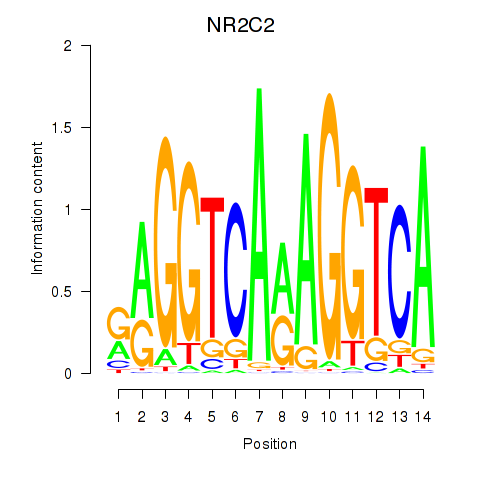
Results for RXRA_NR2F6_NR2C2
Z-value: 0.70
Transcription factors associated with RXRA_NR2F6_NR2C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRA
|
ENSG00000186350.8 | retinoid X receptor alpha |
|
NR2F6
|
ENSG00000160113.5 | nuclear receptor subfamily 2 group F member 6 |
|
NR2C2
|
ENSG00000177463.11 | nuclear receptor subfamily 2 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F6 | hg19_v2_chr19_-_17356697_17356762 | 0.41 | 2.1e-10 | Click! |
| NR2C2 | hg19_v2_chr3_+_14989076_14989113, hg19_v2_chr3_+_14989186_14989236 | -0.37 | 1.7e-08 | Click! |
| RXRA | hg19_v2_chr9_+_137218362_137218426, hg19_v2_chr9_+_137298396_137298431 | -0.10 | 1.4e-01 | Click! |
Activity profile of RXRA_NR2F6_NR2C2 motif
Sorted Z-values of RXRA_NR2F6_NR2C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_132413739 | 30.51 |
ENST00000443358.2
|
PUS1
|
pseudouridylate synthase 1 |
| chr12_+_132413798 | 25.18 |
ENST00000440818.2
ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1
|
pseudouridylate synthase 1 |
| chr12_+_132413765 | 22.84 |
ENST00000376649.3
ENST00000322060.5 |
PUS1
|
pseudouridylate synthase 1 |
| chr8_-_80942061 | 22.47 |
ENST00000519386.1
|
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr19_+_50919056 | 22.21 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr6_+_24775153 | 19.93 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr12_+_96252706 | 19.53 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr3_-_119396193 | 19.40 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr8_-_80942139 | 19.32 |
ENST00000521434.1
ENST00000519120.1 ENST00000520946.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr7_-_72936531 | 17.17 |
ENST00000339594.4
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr17_-_4852332 | 16.12 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr11_-_66206260 | 15.98 |
ENST00000329819.4
ENST00000310999.7 ENST00000430466.2 |
MRPL11
|
mitochondrial ribosomal protein L11 |
| chr6_+_33172407 | 14.76 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr2_+_120125245 | 14.52 |
ENST00000393103.2
|
DBI
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr4_-_7069760 | 13.83 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr8_-_80942467 | 13.00 |
ENST00000518271.1
ENST00000276585.4 ENST00000521605.1 |
MRPS28
|
mitochondrial ribosomal protein S28 |
| chr2_-_219134343 | 12.76 |
ENST00000447885.1
ENST00000420660.1 |
AAMP
|
angio-associated, migratory cell protein |
| chr17_-_4852243 | 12.65 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chr16_+_30075463 | 12.53 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_154947148 | 12.20 |
ENST00000368436.1
ENST00000308987.5 |
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr1_-_193075180 | 12.09 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr2_-_207024134 | 12.03 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr16_+_30075595 | 12.02 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_154947126 | 11.77 |
ENST00000368439.1
|
CKS1B
|
CDC28 protein kinase regulatory subunit 1B |
| chr2_+_200820269 | 11.70 |
ENST00000392290.1
|
C2orf47
|
chromosome 2 open reading frame 47 |
| chr16_+_30075783 | 11.28 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_198365122 | 11.22 |
ENST00000604458.1
|
HSPE1-MOB4
|
HSPE1-MOB4 readthrough |
| chr5_+_1801503 | 10.94 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr1_-_43855444 | 10.86 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr2_-_207024233 | 10.71 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr18_-_54318353 | 10.70 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr19_-_59023348 | 10.65 |
ENST00000601355.1
ENST00000263093.2 |
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr1_+_65613340 | 10.47 |
ENST00000546702.1
|
AK4
|
adenylate kinase 4 |
| chr2_+_86426478 | 10.45 |
ENST00000254644.8
ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35
|
mitochondrial ribosomal protein L35 |
| chr8_+_145582633 | 10.21 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chrX_-_1511617 | 10.04 |
ENST00000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr19_-_17356697 | 9.91 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr1_+_165864800 | 9.37 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_165864821 | 9.30 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr2_+_198365095 | 9.08 |
ENST00000409468.1
|
HSPE1
|
heat shock 10kDa protein 1 |
| chr16_+_2255841 | 8.85 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr16_+_2255710 | 8.61 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr2_+_178257372 | 8.49 |
ENST00000264167.4
ENST00000409888.1 |
AGPS
|
alkylglycerone phosphate synthase |
| chr22_+_30163340 | 8.48 |
ENST00000330029.6
ENST00000401406.3 |
UQCR10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr7_-_65447192 | 8.44 |
ENST00000421103.1
ENST00000345660.6 ENST00000304895.4 |
GUSB
|
glucuronidase, beta |
| chr1_-_6420737 | 8.41 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr11_-_73687997 | 8.35 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr2_-_219134822 | 8.03 |
ENST00000444053.1
ENST00000248450.4 |
AAMP
|
angio-associated, migratory cell protein |
| chr22_-_19165917 | 7.55 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr5_+_271733 | 7.46 |
ENST00000264933.4
|
PDCD6
|
programmed cell death 6 |
| chr19_-_2328572 | 7.35 |
ENST00000252622.10
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_211848899 | 7.27 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chrY_-_1461617 | 7.08 |
ENSTR0000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr12_-_7079805 | 6.79 |
ENST00000536316.2
ENST00000542912.1 ENST00000440277.1 ENST00000545167.1 ENST00000546111.1 ENST00000399433.2 ENST00000535923.1 |
PHB2
|
prohibitin 2 |
| chr6_-_33282163 | 6.55 |
ENST00000434618.2
ENST00000456592.2 |
TAPBP
|
TAP binding protein (tapasin) |
| chr1_-_161193349 | 6.47 |
ENST00000469730.2
ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2
|
apolipoprotein A-II |
| chr5_+_271752 | 6.42 |
ENST00000505221.1
ENST00000509581.1 ENST00000507528.1 |
PDCD6
|
programmed cell death 6 |
| chr16_-_4401258 | 6.39 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr11_+_64073699 | 5.91 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr19_-_51869592 | 5.84 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr15_+_43985084 | 5.81 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr3_-_113465065 | 5.81 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_-_156722015 | 5.75 |
ENST00000368209.5
|
HDGF
|
hepatoma-derived growth factor |
| chr2_+_219283815 | 5.72 |
ENST00000248444.5
ENST00000454069.1 ENST00000392114.2 |
VIL1
|
villin 1 |
| chr11_+_67798363 | 5.70 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr4_+_174089904 | 5.69 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr16_-_4401284 | 5.60 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr17_+_7123125 | 5.54 |
ENST00000356839.5
ENST00000583312.1 ENST00000350303.5 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr2_+_159313452 | 5.53 |
ENST00000389757.3
ENST00000389759.3 |
PKP4
|
plakophilin 4 |
| chr11_+_85339623 | 5.53 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr6_-_33385854 | 5.50 |
ENST00000488478.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr22_-_37415475 | 5.48 |
ENST00000403892.3
ENST00000249042.3 ENST00000438203.1 |
TST
|
thiosulfate sulfurtransferase (rhodanese) |
| chr1_-_156721389 | 5.38 |
ENST00000537739.1
|
HDGF
|
hepatoma-derived growth factor |
| chr2_+_10442993 | 5.34 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr1_+_110527308 | 5.31 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr1_-_8938736 | 5.28 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr15_+_43885252 | 5.20 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr12_+_6977258 | 5.20 |
ENST00000488464.2
ENST00000535434.1 ENST00000493987.1 |
TPI1
|
triosephosphate isomerase 1 |
| chr1_-_155880672 | 5.14 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr3_+_52232102 | 5.14 |
ENST00000469224.1
ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr15_-_76603727 | 5.11 |
ENST00000560595.1
ENST00000433983.2 ENST00000559386.1 ENST00000559602.1 ENST00000560726.1 ENST00000557943.1 |
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr7_-_102232891 | 5.04 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chr1_-_156721502 | 5.02 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr22_-_24316648 | 4.99 |
ENST00000403754.3
ENST00000430101.2 ENST00000398344.4 |
DDT
|
D-dopachrome tautomerase |
| chr7_-_74489609 | 4.97 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr8_-_145331153 | 4.92 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr22_+_24309089 | 4.78 |
ENST00000215770.5
|
DDTL
|
D-dopachrome tautomerase-like |
| chr2_-_74607390 | 4.75 |
ENST00000413111.1
ENST00000409567.3 ENST00000454119.1 ENST00000361874.3 ENST00000394003.3 |
DCTN1
|
dynactin 1 |
| chr3_-_100120223 | 4.72 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr11_+_86013253 | 4.72 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chr12_+_7079944 | 4.71 |
ENST00000261406.6
|
EMG1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr11_+_118443098 | 4.70 |
ENST00000392859.3
ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1
|
archain 1 |
| chr6_-_33385655 | 4.61 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chrX_-_153191708 | 4.60 |
ENST00000393721.1
ENST00000370028.3 |
ARHGAP4
|
Rho GTPase activating protein 4 |
| chrX_+_154611749 | 4.55 |
ENST00000369505.3
|
F8A2
|
coagulation factor VIII-associated 2 |
| chr6_+_13615554 | 4.55 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chrX_-_152760934 | 4.55 |
ENST00000370210.1
ENST00000421080.2 |
HAUS7
|
HAUS augmin-like complex, subunit 7 |
| chr19_-_10687907 | 4.54 |
ENST00000589348.1
|
AP1M2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr11_-_64013663 | 4.53 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr17_-_80017856 | 4.43 |
ENST00000577574.1
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr10_-_96829246 | 4.39 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr1_-_36930012 | 4.38 |
ENST00000373116.5
|
MRPS15
|
mitochondrial ribosomal protein S15 |
| chr11_-_6640585 | 4.34 |
ENST00000533371.1
ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1
|
tripeptidyl peptidase I |
| chr6_+_143772060 | 4.31 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr5_+_162864575 | 4.26 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr17_-_61920280 | 4.24 |
ENST00000448276.2
ENST00000577990.1 |
SMARCD2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr6_-_31926629 | 4.23 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr13_+_50656307 | 4.23 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr6_-_33385870 | 4.18 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr11_+_46383121 | 4.13 |
ENST00000454345.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr17_+_37894179 | 4.10 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chr14_-_75643296 | 4.07 |
ENST00000303575.4
|
TMED10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr3_-_197300194 | 4.06 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr11_+_67798090 | 4.04 |
ENST00000313468.5
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chrX_-_153285251 | 3.95 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr16_+_4475806 | 3.88 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr22_+_20105012 | 3.87 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr1_+_113161778 | 3.81 |
ENST00000263168.3
|
CAPZA1
|
capping protein (actin filament) muscle Z-line, alpha 1 |
| chr6_-_33385823 | 3.81 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr17_+_46970178 | 3.74 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_46970134 | 3.69 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chrX_-_153285395 | 3.68 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr1_+_26869597 | 3.68 |
ENST00000530003.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr6_-_33385902 | 3.67 |
ENST00000374500.5
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr12_-_56694142 | 3.66 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr17_+_42427826 | 3.62 |
ENST00000586443.1
|
GRN
|
granulin |
| chr16_-_67970990 | 3.60 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr6_-_31704282 | 3.55 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr7_-_107204918 | 3.55 |
ENST00000297135.3
|
COG5
|
component of oligomeric golgi complex 5 |
| chr5_-_68665296 | 3.54 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr6_+_88299833 | 3.52 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr17_-_79895097 | 3.50 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr17_+_46970127 | 3.48 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr1_+_38478378 | 3.37 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr4_-_103266626 | 3.36 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr11_+_67798114 | 3.35 |
ENST00000453471.2
ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr19_+_11546093 | 3.34 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr17_-_79894651 | 3.34 |
ENST00000584848.1
ENST00000577756.1 ENST00000329875.8 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr17_-_27503770 | 3.32 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr8_+_22224811 | 3.31 |
ENST00000381237.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr21_-_45196326 | 3.30 |
ENST00000291568.5
|
CSTB
|
cystatin B (stefin B) |
| chr19_+_535835 | 3.30 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr5_-_68665815 | 3.29 |
ENST00000380818.3
ENST00000328663.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr17_-_42144949 | 3.26 |
ENST00000591247.1
|
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr11_-_57298187 | 3.25 |
ENST00000525158.1
ENST00000257245.4 ENST00000525587.1 |
TIMM10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr8_+_22224760 | 3.23 |
ENST00000359741.5
ENST00000520644.1 ENST00000240095.6 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr19_+_10362882 | 3.18 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_-_155271213 | 3.17 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr1_+_145727681 | 3.14 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr22_+_37415676 | 3.14 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr17_+_41150793 | 3.09 |
ENST00000586277.1
|
RPL27
|
ribosomal protein L27 |
| chr16_-_47007545 | 3.08 |
ENST00000317089.5
|
DNAJA2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr19_+_10197463 | 3.08 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr15_-_82555000 | 3.05 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr22_+_44351419 | 3.03 |
ENST00000396202.3
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr3_-_47517302 | 3.02 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr1_-_155270770 | 3.00 |
ENST00000392414.3
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr22_+_37415728 | 3.00 |
ENST00000404802.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr20_-_48729670 | 3.00 |
ENST00000371657.5
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr22_+_37415700 | 2.98 |
ENST00000397129.1
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr11_+_64001962 | 2.98 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr1_+_151129135 | 2.96 |
ENST00000602841.1
|
SCNM1
|
sodium channel modifier 1 |
| chr22_+_37415776 | 2.95 |
ENST00000341116.3
ENST00000429360.2 ENST00000404393.1 |
MPST
|
mercaptopyruvate sulfurtransferase |
| chr17_+_1646130 | 2.93 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chrX_-_70474910 | 2.91 |
ENST00000373988.1
ENST00000373998.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr17_+_41150479 | 2.88 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr12_-_6960407 | 2.85 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr17_+_41150290 | 2.80 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chrX_-_15511438 | 2.78 |
ENST00000380420.5
|
PIR
|
pirin (iron-binding nuclear protein) |
| chr12_+_52463751 | 2.77 |
ENST00000336854.4
ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44
|
chromosome 12 open reading frame 44 |
| chr18_-_47340297 | 2.75 |
ENST00000586485.1
ENST00000587994.1 ENST00000586100.1 ENST00000285093.10 |
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr22_+_44351301 | 2.75 |
ENST00000350028.4
|
SAMM50
|
SAMM50 sorting and assembly machinery component |
| chr2_+_28113583 | 2.71 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr5_-_68665469 | 2.70 |
ENST00000217893.5
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr11_+_71938925 | 2.70 |
ENST00000538751.1
|
INPPL1
|
inositol polyphosphate phosphatase-like 1 |
| chr19_+_11546440 | 2.69 |
ENST00000589126.1
ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH
|
protein kinase C substrate 80K-H |
| chr12_+_67663056 | 2.67 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr17_+_37821593 | 2.64 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr5_-_68665084 | 2.63 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr1_-_173886491 | 2.63 |
ENST00000367698.3
|
SERPINC1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr11_+_64002292 | 2.62 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr22_+_20105259 | 2.56 |
ENST00000416427.1
ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1
|
RAN binding protein 1 |
| chr2_+_234621551 | 2.53 |
ENST00000608381.1
ENST00000373414.3 |
UGT1A1
UGT1A5
|
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr2_+_217498105 | 2.52 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr1_-_109968973 | 2.51 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr3_+_113465866 | 2.50 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr15_+_43985725 | 2.39 |
ENST00000413453.2
|
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr19_-_51466681 | 2.37 |
ENST00000456750.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr3_-_11610255 | 2.36 |
ENST00000424529.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr19_+_45394477 | 2.36 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr19_-_8008533 | 2.36 |
ENST00000597926.1
|
TIMM44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr1_+_110163202 | 2.34 |
ENST00000531203.1
ENST00000256578.3 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr17_+_75446819 | 2.33 |
ENST00000541152.2
ENST00000591704.1 |
SEPT9
|
septin 9 |
| chr22_+_23077065 | 2.32 |
ENST00000390310.2
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chrX_-_41782249 | 2.27 |
ENST00000442742.2
ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr17_+_7155556 | 2.24 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr5_+_156712372 | 2.23 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr14_-_25519095 | 2.21 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr19_+_44100632 | 2.21 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr11_+_394196 | 2.21 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRA_NR2F6_NR2C2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.2 | 78.5 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 5.7 | 17.1 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 4.7 | 18.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 3.5 | 3.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 3.5 | 17.5 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 3.4 | 10.3 | GO:2000909 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 3.3 | 19.9 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 2.9 | 28.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 2.8 | 8.5 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 2.6 | 10.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 2.4 | 14.5 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 2.3 | 13.9 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 2.1 | 8.4 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 2.0 | 30.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 2.0 | 35.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.9 | 7.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 1.7 | 5.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 1.5 | 10.6 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.5 | 7.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 1.3 | 12.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 1.2 | 1.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 1.2 | 12.1 | GO:0042262 | DNA protection(GO:0042262) |
| 1.2 | 88.8 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 1.2 | 14.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.2 | 4.7 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 1.1 | 11.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 1.1 | 5.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 1.1 | 6.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 1.1 | 3.3 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 1.1 | 8.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 1.0 | 4.1 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 1.0 | 6.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 1.0 | 4.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 1.0 | 6.8 | GO:0033600 | positive regulation of exit from mitosis(GO:0031536) negative regulation of mammary gland epithelial cell proliferation(GO:0033600) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.0 | 5.8 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.9 | 12.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.9 | 5.5 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.9 | 14.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.9 | 2.6 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.8 | 8.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.8 | 4.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.8 | 4.7 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.8 | 7.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.8 | 59.1 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.8 | 11.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.8 | 5.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.7 | 8.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.7 | 4.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.7 | 5.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.7 | 3.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 6.4 | GO:1990668 | vesicle fusion with endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane(GO:1990668) |
| 0.6 | 4.4 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.6 | 0.6 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.6 | 3.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.6 | 4.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.6 | 4.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.6 | 19.5 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.6 | 0.6 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.5 | 17.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.5 | 1.6 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.5 | 3.7 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.5 | 1.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.5 | 4.7 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.5 | 6.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.5 | 24.0 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.5 | 5.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.5 | 1.4 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.5 | 3.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.5 | 1.4 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.4 | 8.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.4 | 2.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 3.9 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.4 | 2.9 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.4 | 3.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.4 | 2.5 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.4 | 2.0 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.4 | 10.1 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.4 | 1.2 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.4 | 5.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.4 | 7.6 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.4 | 10.9 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 8.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.3 | 2.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 0.9 | GO:2000395 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.3 | 1.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.3 | 4.2 | GO:1900364 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.3 | 0.6 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.3 | 0.9 | GO:0050894 | determination of affect(GO:0050894) |
| 0.3 | 0.6 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.3 | 3.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) positive regulation of inclusion body assembly(GO:0090261) |
| 0.3 | 0.8 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.3 | 2.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 7.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.3 | 20.8 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.2 | 1.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 0.9 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.2 | 1.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.2 | 2.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.2 | 0.6 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 3.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.8 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 3.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.2 | 0.8 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) |
| 0.2 | 0.8 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 0.9 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.2 | 4.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 4.3 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 3.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.2 | 16.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 1.5 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 2.5 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 0.7 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 1.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 2.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 2.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 5.0 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 1.1 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body biosynthetic process(GO:0046951) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 1.6 | GO:0051873 | killing by host of symbiont cells(GO:0051873) |
| 0.1 | 4.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 3.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 4.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.4 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.1 | 3.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 3.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 5.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 0.2 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 6.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 3.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 3.1 | GO:0090314 | positive regulation of protein targeting to membrane(GO:0090314) |
| 0.1 | 1.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.2 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.1 | 1.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 4.8 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.4 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 5.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 1.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 2.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.4 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.3 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.1 | 2.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 2.5 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.9 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.1 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.7 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 7.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 0.5 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 | 1.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.1 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 5.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 4.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 1.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.5 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.1 | 1.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 8.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.9 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 1.0 | GO:0019184 | glutathione biosynthetic process(GO:0006750) nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.1 | 3.6 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 2.8 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 1.0 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 0.8 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.1 | 0.4 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.1 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.3 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 4.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 1.6 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.8 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 2.2 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.3 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.4 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.5 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.7 | GO:0070873 | regulation of polysaccharide metabolic process(GO:0032881) regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.6 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 1.4 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 13.0 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 2.3 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 0.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 3.9 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 25.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 4.5 | 22.7 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 2.5 | 20.4 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 2.4 | 19.5 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 2.2 | 18.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 2.2 | 59.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 1.5 | 7.6 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 1.5 | 12.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 1.2 | 3.5 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 1.1 | 6.4 | GO:0042825 | TAP complex(GO:0042825) |
| 1.1 | 7.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 1.0 | 5.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.9 | 5.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.8 | 4.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.8 | 5.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.8 | 29.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.8 | 8.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.7 | 8.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.7 | 16.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.7 | 20.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.6 | 10.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.6 | 10.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 1.8 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.6 | 5.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.6 | 29.5 | GO:0031430 | M band(GO:0031430) |
| 0.5 | 3.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 14.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.5 | 4.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.5 | 4.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 3.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 2.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 24.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.4 | 25.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 4.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 20.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.3 | 145.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.3 | 4.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.3 | 4.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 11.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 0.8 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.3 | 1.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 4.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 5.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 7.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 2.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 10.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.8 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 2.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 0.9 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 4.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 3.0 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 2.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 8.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 4.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.2 | 12.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 3.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 32.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 3.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 4.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 4.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 29.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 11.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 1.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 10.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 6.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 8.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 22.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 4.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 3.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 3.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 13.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 2.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.5 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 2.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.7 | GO:0032994 | protein-lipid complex(GO:0032994) |
| 0.0 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 3.4 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 1.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.8 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 26.2 | 78.5 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 4.3 | 17.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 3.6 | 14.5 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 3.6 | 42.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 3.4 | 24.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 3.3 | 35.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 3.2 | 19.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 3.0 | 12.1 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 3.0 | 14.8 | GO:0070404 | NADH binding(GO:0070404) |
| 2.7 | 5.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 2.7 | 10.9 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 2.7 | 18.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 2.6 | 10.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 2.4 | 12.1 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 2.3 | 22.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 2.1 | 8.4 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 1.9 | 7.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 1.7 | 5.1 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 1.7 | 6.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 1.6 | 6.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 1.5 | 6.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.5 | 10.7 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 1.4 | 5.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 1.4 | 4.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 1.2 | 5.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 1.2 | 22.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 1.2 | 8.5 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.2 | 7.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.1 | 7.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 1.1 | 7.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.0 | 8.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.9 | 4.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.9 | 5.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.9 | 5.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.9 | 4.4 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.9 | 12.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.8 | 5.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.8 | 3.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.8 | 12.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.7 | 4.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.7 | 5.7 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.7 | 9.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.7 | 12.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.6 | 9.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.6 | 12.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 2.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.5 | 17.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.5 | 14.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.5 | 2.6 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.5 | 25.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.5 | 1.6 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.5 | 5.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 1.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.4 | 5.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 0.4 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.4 | 10.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 0.4 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.4 | 7.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.4 | 5.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.4 | 3.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 1.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.4 | 8.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 4.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.3 | 0.9 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.3 | 2.8 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 2.7 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.3 | 0.9 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.3 | 4.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 1.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.3 | 2.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 4.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 0.7 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 17.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 1.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 2.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.2 | 4.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 2.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 32.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 15.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.2 | 1.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 3.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.8 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.2 | 6.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 2.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.2 | 17.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 2.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 1.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 4.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 26.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.1 | 2.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 3.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 3.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 4.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 7.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 4.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 10.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 2.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 2.7 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 2.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 4.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 3.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 5.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 2.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 3.3 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.4 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 6.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 5.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.7 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.6 | GO:0098847 | sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.1 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 5.9 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 5.1 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 36.6 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 3.1 | GO:0016829 | lyase activity(GO:0016829) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 5.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.9 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.3 | GO:0015923 | alpha-mannosidase activity(GO:0004559) mannosidase activity(GO:0015923) |
| 0.0 | 1.4 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 41.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.4 | 31.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.4 | 28.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.4 | 5.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 9.3 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.3 | 17.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 12.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.3 | 14.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.3 | 15.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 10.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 3.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 4.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 4.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 3.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 3.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 6.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 95.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 1.2 | 23.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 1.2 | 19.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 1.1 | 10.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 1.0 | 51.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.9 | 9.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.8 | 17.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 18.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.5 | 4.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 7.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.4 | 8.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 24.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.4 | 36.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.4 | 6.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 8.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 29.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.4 | 19.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.4 | 7.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 7.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.3 | 7.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.3 | 2.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.3 | 7.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 7.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 6.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 7.3 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.2 | 9.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 4.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 7.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 0.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.2 | 12.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 1.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 16.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 2.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 2.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 3.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 4.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 6.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 2.9 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 7.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.7 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 7.2 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.1 | 2.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 7.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 8.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 6.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 6.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |