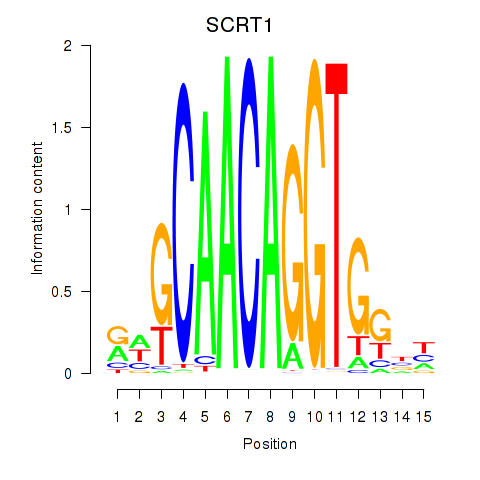
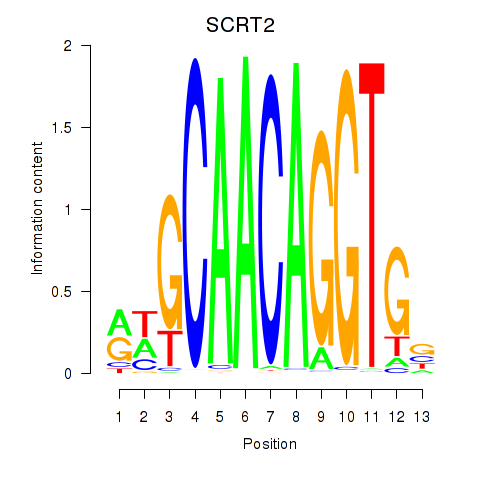
Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
Results for SCRT1_SCRT2
Z-value: 0.36
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | -0.27 | 7.0e-05 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41258786 | 59.22 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr6_+_121756809 | 55.40 |
ENST00000282561.3
|
GJA1
|
gap junction protein, alpha 1, 43kDa |
| chr15_-_72523924 | 34.00 |
ENST00000566809.1
ENST00000567087.1 ENST00000569050.1 ENST00000568883.1 |
PKM
|
pyruvate kinase, muscle |
| chr15_-_72523454 | 32.85 |
ENST00000565154.1
ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM
|
pyruvate kinase, muscle |
| chr1_-_95392635 | 28.72 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr15_-_30113676 | 25.42 |
ENST00000400011.2
|
TJP1
|
tight junction protein 1 |
| chr2_-_175870085 | 25.41 |
ENST00000409156.3
|
CHN1
|
chimerin 1 |
| chr5_-_73936451 | 23.83 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chrX_+_51636629 | 23.01 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr15_-_30114231 | 21.47 |
ENST00000356107.6
ENST00000545208.2 |
TJP1
|
tight junction protein 1 |
| chr14_+_23775971 | 21.44 |
ENST00000250405.5
|
BCL2L2
|
BCL2-like 2 |
| chr4_+_41540160 | 20.11 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_33283754 | 18.19 |
ENST00000373477.4
|
YARS
|
tyrosyl-tRNA synthetase |
| chr11_-_117748138 | 17.50 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr1_-_222885770 | 14.32 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr11_-_117747607 | 14.32 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr20_-_43977055 | 14.25 |
ENST00000372733.3
ENST00000537976.1 |
SDC4
|
syndecan 4 |
| chr11_-_117747434 | 14.00 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr10_+_3109695 | 11.99 |
ENST00000381125.4
|
PFKP
|
phosphofructokinase, platelet |
| chr1_+_16083154 | 11.99 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr11_+_32112431 | 11.65 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr8_+_98881268 | 11.27 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chrX_+_118370211 | 10.65 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr19_-_46974664 | 10.65 |
ENST00000438932.2
|
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr19_-_46974741 | 10.52 |
ENST00000313683.10
ENST00000602246.1 |
PNMAL1
|
paraneoplastic Ma antigen family-like 1 |
| chr5_+_125758813 | 10.28 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758865 | 10.13 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr3_-_42846021 | 10.09 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr9_+_114423615 | 10.03 |
ENST00000374293.4
|
GNG10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr3_+_167453493 | 9.87 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr12_+_53342625 | 9.52 |
ENST00000388837.2
ENST00000550600.1 ENST00000388835.3 |
KRT18
|
keratin 18 |
| chr6_-_83903600 | 9.18 |
ENST00000506587.1
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr19_+_40877583 | 9.02 |
ENST00000596470.1
|
PLD3
|
phospholipase D family, member 3 |
| chrX_+_51927919 | 8.86 |
ENST00000416960.1
|
MAGED4
|
melanoma antigen family D, 4 |
| chr9_+_125703282 | 8.83 |
ENST00000373647.4
ENST00000402311.1 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr14_+_23776024 | 8.64 |
ENST00000553781.1
ENST00000556100.1 ENST00000557236.1 ENST00000557579.1 |
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough BCL2-like 2 |
| chrX_-_51812268 | 8.50 |
ENST00000486010.1
ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B
|
melanoma antigen family D, 4B |
| chr7_+_130126165 | 8.10 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr4_-_5890145 | 7.99 |
ENST00000397890.2
|
CRMP1
|
collapsin response mediator protein 1 |
| chr3_+_133465228 | 7.89 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr2_-_224467093 | 7.67 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr14_+_23776167 | 7.62 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr8_+_132952112 | 7.60 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr7_+_130126012 | 7.51 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr1_+_225965518 | 7.42 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr12_-_21810765 | 7.41 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr16_-_53537105 | 7.39 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr21_-_35987438 | 7.38 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr11_+_69455855 | 7.37 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr1_-_94312706 | 7.36 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_-_21810726 | 7.34 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr11_-_111794446 | 7.34 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr5_+_74011328 | 7.26 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr16_+_2039946 | 7.22 |
ENST00000248121.2
ENST00000568896.1 |
SYNGR3
|
synaptogyrin 3 |
| chrX_+_51928002 | 6.80 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr16_-_50402690 | 6.79 |
ENST00000394689.2
|
BRD7
|
bromodomain containing 7 |
| chr3_-_42845951 | 6.75 |
ENST00000418900.2
ENST00000430190.1 |
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr4_-_140223670 | 6.74 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_-_73150599 | 6.66 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr8_+_22429205 | 5.90 |
ENST00000520207.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr3_-_79816965 | 5.81 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_+_138089100 | 5.73 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr2_+_187350883 | 5.63 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr1_-_156308018 | 5.62 |
ENST00000496684.2
ENST00000368259.2 ENST00000368261.3 ENST00000472765.2 ENST00000533194.1 ENST00000478640.2 |
CCT3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr6_-_10419871 | 5.57 |
ENST00000319516.4
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr11_+_61583721 | 5.24 |
ENST00000257261.6
|
FADS2
|
fatty acid desaturase 2 |
| chr7_-_99698338 | 5.18 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr17_-_40288449 | 5.13 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr20_+_57427765 | 5.12 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr19_-_58609570 | 5.09 |
ENST00000600845.1
ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18
|
zinc finger and SCAN domain containing 18 |
| chr17_-_1083078 | 4.90 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr16_+_103816 | 4.90 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr3_+_141144954 | 4.88 |
ENST00000441582.2
ENST00000321464.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_+_37423618 | 4.77 |
ENST00000402297.1
ENST00000397064.2 ENST00000406711.1 ENST00000392061.2 ENST00000397226.2 |
AC007390.5
|
CEBPZ antisense RNA 1 |
| chr15_+_43985084 | 4.75 |
ENST00000434505.1
ENST00000411750.1 |
CKMT1A
|
creatine kinase, mitochondrial 1A |
| chr3_-_62861012 | 4.73 |
ENST00000357948.3
ENST00000383710.4 |
CADPS
|
Ca++-dependent secretion activator |
| chr15_+_43885252 | 4.62 |
ENST00000453782.1
ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chrX_-_153141302 | 4.54 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_+_30097181 | 4.47 |
ENST00000586420.1
ENST00000221770.3 ENST00000392279.3 ENST00000590688.1 |
POP4
|
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr5_-_111091948 | 4.36 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr11_-_71159380 | 4.02 |
ENST00000525346.1
ENST00000531364.1 ENST00000529990.1 ENST00000527316.1 ENST00000407721.2 |
DHCR7
|
7-dehydrocholesterol reductase |
| chr2_+_187350973 | 3.91 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr17_+_53343577 | 3.88 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr22_+_40742497 | 3.82 |
ENST00000216194.7
|
ADSL
|
adenylosuccinate lyase |
| chr14_+_67708137 | 3.77 |
ENST00000556345.1
ENST00000555925.1 ENST00000557783.1 |
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr22_+_40742512 | 3.65 |
ENST00000454266.2
ENST00000342312.6 |
ADSL
|
adenylosuccinate lyase |
| chr10_-_47222824 | 3.65 |
ENST00000355232.3
|
AGAP10
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr14_+_67707826 | 3.59 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr2_+_173600671 | 3.57 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_-_52456352 | 3.47 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr19_+_35739897 | 3.47 |
ENST00000605618.1
ENST00000427250.1 ENST00000601623.1 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr11_+_64002292 | 3.47 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr6_-_31697255 | 3.42 |
ENST00000436437.1
|
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr14_-_102829051 | 3.37 |
ENST00000536961.2
ENST00000541568.2 ENST00000216756.6 |
CINP
|
cyclin-dependent kinase 2 interacting protein |
| chr2_+_173600565 | 3.27 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_71107921 | 3.23 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr7_+_12727250 | 3.22 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr19_+_35739782 | 3.21 |
ENST00000347609.4
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr3_+_87276407 | 3.21 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr6_+_69345166 | 3.20 |
ENST00000370598.1
|
BAI3
|
brain-specific angiogenesis inhibitor 3 |
| chr1_+_109289279 | 3.18 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr16_+_25123041 | 3.17 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr2_+_173600514 | 3.13 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr4_-_90757364 | 3.04 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_+_842808 | 2.97 |
ENST00000397397.2
ENST00000397411.2 ENST00000397396.1 |
TSPAN4
|
tetraspanin 4 |
| chr5_+_173763250 | 2.96 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr11_+_842928 | 2.94 |
ENST00000397408.1
|
TSPAN4
|
tetraspanin 4 |
| chr4_-_52904425 | 2.92 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr12_-_71148357 | 2.91 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr11_+_27062272 | 2.89 |
ENST00000529202.1
ENST00000533566.1 |
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr7_-_122526499 | 2.78 |
ENST00000412584.2
|
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr17_-_43025005 | 2.66 |
ENST00000587309.1
ENST00000593135.1 ENST00000339151.4 |
KIF18B
|
kinesin family member 18B |
| chr22_-_19466683 | 2.56 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr17_-_32484313 | 2.40 |
ENST00000359872.6
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr22_-_19466732 | 2.38 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr13_+_58206655 | 2.34 |
ENST00000377918.3
|
PCDH17
|
protocadherin 17 |
| chr4_+_165675269 | 2.31 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr12_-_52911718 | 2.26 |
ENST00000548409.1
|
KRT5
|
keratin 5 |
| chrX_+_13707235 | 2.23 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr16_-_103572 | 2.21 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr1_-_51425902 | 2.19 |
ENST00000396153.2
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr11_-_61659006 | 2.19 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr10_+_35484793 | 2.18 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr8_-_116681221 | 2.15 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_-_31697563 | 2.14 |
ENST00000375789.2
ENST00000416410.1 |
DDAH2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_15500280 | 2.10 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr10_+_35484053 | 2.03 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr12_-_13153139 | 2.00 |
ENST00000536942.1
ENST00000014930.4 |
HEBP1
|
heme binding protein 1 |
| chr20_-_45061695 | 1.99 |
ENST00000445496.2
|
ELMO2
|
engulfment and cell motility 2 |
| chr18_+_47088401 | 1.96 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr2_+_223289208 | 1.95 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr12_-_71148413 | 1.89 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_224033596 | 1.88 |
ENST00000391878.2
ENST00000343537.7 |
TP53BP2
|
tumor protein p53 binding protein, 2 |
| chr17_-_62340581 | 1.81 |
ENST00000258991.3
ENST00000583738.1 ENST00000584379.1 |
TEX2
|
testis expressed 2 |
| chr7_-_122526799 | 1.78 |
ENST00000334010.7
ENST00000313070.7 |
CADPS2
|
Ca++-dependent secretion activator 2 |
| chr3_-_196910721 | 1.78 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_-_156918806 | 1.76 |
ENST00000315174.8
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr12_-_122907091 | 1.74 |
ENST00000358808.2
ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr2_-_241759622 | 1.70 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr1_-_51425772 | 1.62 |
ENST00000371778.4
|
FAF1
|
Fas (TNFRSF6) associated factor 1 |
| chr16_-_30022293 | 1.56 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr1_-_9129895 | 1.55 |
ENST00000473209.1
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chrX_+_70503037 | 1.48 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr14_+_102829300 | 1.46 |
ENST00000359520.7
|
TECPR2
|
tectonin beta-propeller repeat containing 2 |
| chr1_+_150245099 | 1.43 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr4_-_119757322 | 1.43 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D |
| chr19_+_35739631 | 1.42 |
ENST00000602003.1
ENST00000360798.3 ENST00000354900.3 |
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr1_-_212588157 | 1.37 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr19_-_5838768 | 1.34 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr11_+_6624955 | 1.32 |
ENST00000299421.4
ENST00000537806.1 |
ILK
|
integrin-linked kinase |
| chr21_-_34852304 | 1.30 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr6_+_90272027 | 1.25 |
ENST00000522441.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr19_+_35739597 | 1.25 |
ENST00000361790.3
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr3_-_97690931 | 1.21 |
ENST00000360258.4
|
MINA
|
MYC induced nuclear antigen |
| chr12_-_57824739 | 1.18 |
ENST00000347140.3
ENST00000402412.1 |
R3HDM2
|
R3H domain containing 2 |
| chr14_+_105886150 | 1.17 |
ENST00000331320.7
ENST00000406191.1 |
MTA1
|
metastasis associated 1 |
| chr3_-_186080012 | 1.15 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr11_-_67169253 | 1.14 |
ENST00000527663.1
ENST00000312989.7 |
PPP1CA
|
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr1_-_153940097 | 1.13 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr16_+_2198604 | 1.09 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr12_+_101988627 | 0.99 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr4_-_83483395 | 0.97 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr17_+_63133526 | 0.97 |
ENST00000443584.3
|
RGS9
|
regulator of G-protein signaling 9 |
| chr15_+_80351910 | 0.97 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr12_-_75905374 | 0.95 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr4_-_141348789 | 0.95 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr19_+_35739280 | 0.93 |
ENST00000602122.1
|
LSR
|
lipolysis stimulated lipoprotein receptor |
| chr1_+_110162448 | 0.92 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chrX_-_31285042 | 0.92 |
ENST00000378680.2
ENST00000378723.3 |
DMD
|
dystrophin |
| chr17_-_79805146 | 0.85 |
ENST00000415593.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr3_-_114343768 | 0.84 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr5_+_140430979 | 0.84 |
ENST00000306549.3
|
PCDHB1
|
protocadherin beta 1 |
| chr11_+_6624970 | 0.80 |
ENST00000420936.2
ENST00000528995.1 |
ILK
|
integrin-linked kinase |
| chr15_+_84904525 | 0.73 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr1_-_9129631 | 0.71 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr4_-_119757239 | 0.69 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr1_-_204135450 | 0.62 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr6_-_49755019 | 0.56 |
ENST00000304801.3
|
PGK2
|
phosphoglycerate kinase 2 |
| chr5_-_147211226 | 0.49 |
ENST00000296695.5
|
SPINK1
|
serine peptidase inhibitor, Kazal type 1 |
| chr8_+_145438870 | 0.49 |
ENST00000527931.1
|
FAM203B
|
family with sequence similarity 203, member B |
| chr2_+_61244697 | 0.46 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr17_-_10101868 | 0.46 |
ENST00000432992.2
ENST00000540214.1 |
GAS7
|
growth arrest-specific 7 |
| chr5_+_32710736 | 0.42 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr9_+_99212403 | 0.40 |
ENST00000375251.3
ENST00000375249.4 |
HABP4
|
hyaluronan binding protein 4 |
| chr4_+_89378261 | 0.32 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr10_-_101945771 | 0.30 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr9_+_100174344 | 0.26 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr10_-_95209 | 0.24 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr6_+_71122974 | 0.23 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr19_-_3063099 | 0.20 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chr19_+_12862604 | 0.02 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.7 | 59.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 15.6 | 46.9 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 13.9 | 55.4 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 11.1 | 66.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 5.4 | 37.7 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 2.8 | 14.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 2.7 | 8.0 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 2.6 | 23.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 2.5 | 7.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 2.5 | 7.4 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 2.1 | 10.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 1.9 | 5.8 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 1.9 | 5.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.9 | 9.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 1.8 | 9.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 1.6 | 4.9 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 1.5 | 7.3 | GO:0007619 | courtship behavior(GO:0007619) |
| 1.4 | 5.6 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 1.3 | 7.9 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 1.2 | 28.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 1.1 | 14.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 1.0 | 14.8 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.9 | 12.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.9 | 23.0 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.8 | 7.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.8 | 4.0 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.8 | 4.8 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.8 | 12.0 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.8 | 25.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.7 | 7.4 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.7 | 5.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.7 | 3.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.6 | 3.0 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.6 | 11.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.5 | 5.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 1.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.5 | 2.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.5 | 7.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 1.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.4 | 5.6 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 4.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.4 | 30.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.4 | 3.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.4 | 5.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 5.6 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 2.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.3 | 2.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.3 | 2.9 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.3 | 2.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.3 | 3.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.3 | 18.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.3 | 10.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.3 | 7.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.3 | 5.2 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.3 | 2.9 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.3 | 1.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.3 | 0.9 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.3 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 | 1.8 | GO:1903760 | regulation of potassium ion import(GO:1903286) regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.2 | 4.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 3.8 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 2.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 7.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.2 | 7.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 10.0 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.2 | 11.3 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.2 | 3.2 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 6.8 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 1.9 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 0.6 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.2 | 0.5 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.2 | 6.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 16.8 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.1 | 2.0 | GO:0006670 | sphingosine metabolic process(GO:0006670) diol metabolic process(GO:0034311) |
| 0.1 | 8.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 9.0 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.1 | 7.2 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 6.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.3 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 0.3 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 5.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 2.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 3.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 12.9 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.1 | 6.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.2 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 16.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 2.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 7.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 3.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 4.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.1 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 2.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 2.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 1.0 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.4 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 2.2 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 9.0 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 2.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 4.9 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 2.0 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 1.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.9 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 3.4 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.6 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 7.4 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 55.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 3.6 | 18.2 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 2.9 | 37.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.8 | 7.4 | GO:0070695 | FHF complex(GO:0070695) |
| 1.7 | 10.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 1.6 | 46.9 | GO:0005921 | gap junction(GO:0005921) |
| 1.3 | 7.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 1.3 | 60.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 1.1 | 7.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.8 | 5.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.8 | 9.1 | GO:0043219 | lateral loop(GO:0043219) |
| 0.6 | 4.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.6 | 1.7 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.5 | 7.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 5.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.4 | 7.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 5.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 3.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.4 | 2.7 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.4 | 2.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 39.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.3 | 23.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.3 | 77.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.3 | 3.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.3 | 14.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 22.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.2 | 10.1 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 4.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 4.2 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 4.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 12.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 2.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 7.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 8.8 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 11.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 5.1 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 7.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 7.4 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.1 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 7.3 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 8.0 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.6 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 3.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 2.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 3.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 15.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 4.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 8.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.5 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 26.9 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 6.8 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.4 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.7 | 66.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 14.8 | 59.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 9.2 | 55.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 2.6 | 7.9 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 2.5 | 7.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 2.3 | 9.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 2.1 | 14.8 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 2.0 | 12.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 1.9 | 37.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 1.7 | 5.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 1.5 | 9.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.5 | 39.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 1.4 | 14.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 1.2 | 7.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.1 | 18.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 1.1 | 5.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.0 | 5.8 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 1.0 | 20.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.9 | 7.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.9 | 7.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.7 | 2.9 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.6 | 3.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.6 | 33.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.6 | 3.0 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 4.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.6 | 3.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 5.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.5 | 25.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.5 | 2.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.5 | 5.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 5.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 4.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 4.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 1.3 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.3 | 7.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 7.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 7.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 6.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 0.8 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 46.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.2 | 4.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 6.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 7.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 3.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 5.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 6.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 5.2 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 3.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 5.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 9.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 12.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 2.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 4.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 8.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 25.8 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 8.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.1 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 3.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 14.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 15.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 9.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 7.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 8.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 5.0 | GO:0019904 | protein domain specific binding(GO:0019904) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 56.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 1.1 | 61.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 1.0 | 62.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.4 | 22.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.3 | 14.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 9.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.3 | 23.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 7.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 11.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 6.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.2 | 15.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 3.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 39.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 2.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 5.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 11.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 5.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 4.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.8 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 10.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 55.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 2.5 | 46.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 1.8 | 23.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.6 | 9.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.6 | 15.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.5 | 14.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.5 | 18.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.5 | 14.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 12.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.5 | 7.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 7.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 5.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 7.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 8.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 5.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 5.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 3.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 7.9 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 12.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 10.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 5.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 32.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 5.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 4.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 4.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 5.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 9.0 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 6.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 5.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 2.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 7.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 3.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 8.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |