Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
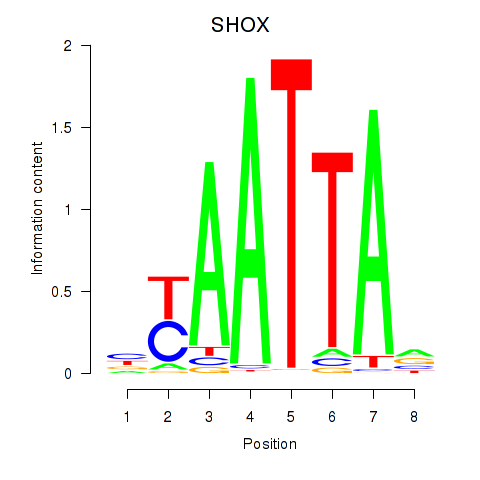
Results for SHOX
Z-value: 0.33
Transcription factors associated with SHOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX
|
ENSG00000185960.8 | short stature homeobox |
|
SHOX
|
ENSGR0000185960.8 | short stature homeobox |
Activity profile of SHOX motif
Sorted Z-values of SHOX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_117748138 | 23.84 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117747607 | 23.45 |
ENST00000540359.1
ENST00000539526.1 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chrX_-_13835147 | 23.17 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr11_-_117747434 | 23.07 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_111794446 | 14.61 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr12_-_16761007 | 13.03 |
ENST00000354662.1
ENST00000441439.2 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr3_+_111718036 | 9.34 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr3_+_111718173 | 9.33 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr13_-_36429763 | 8.92 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr3_+_111717600 | 8.80 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr3_+_111717511 | 8.57 |
ENST00000478951.1
ENST00000393917.2 |
TAGLN3
|
transgelin 3 |
| chr3_+_115342349 | 8.18 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr12_-_16759711 | 7.87 |
ENST00000447609.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_57547870 | 7.78 |
ENST00000381260.3
ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX
|
HOP homeobox |
| chr4_-_57547454 | 7.40 |
ENST00000556376.2
|
HOPX
|
HOP homeobox |
| chr2_-_224467093 | 6.04 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr17_-_9929581 | 5.52 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr5_-_24645078 | 5.50 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr11_-_129062093 | 5.45 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr12_-_10282836 | 5.29 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr18_-_21977748 | 5.16 |
ENST00000399441.4
ENST00000319481.3 |
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr12_-_16758059 | 5.03 |
ENST00000261169.6
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr17_-_78450398 | 4.84 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr7_-_37488834 | 4.53 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_-_102803163 | 4.34 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr1_-_45956822 | 4.32 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr13_-_67802549 | 4.31 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_+_153747746 | 4.27 |
ENST00000368661.3
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr16_-_29910853 | 4.23 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr1_+_66458072 | 4.19 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr10_+_111765562 | 4.15 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr19_+_50016610 | 3.61 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_-_39693111 | 3.57 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr16_-_4852915 | 3.56 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr1_+_180601139 | 3.55 |
ENST00000367590.4
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr1_+_153746683 | 3.53 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr7_-_137028534 | 3.50 |
ENST00000348225.2
|
PTN
|
pleiotrophin |
| chr7_+_154002189 | 3.41 |
ENST00000332007.3
|
DPP6
|
dipeptidyl-peptidase 6 |
| chr12_+_26348246 | 3.40 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr8_+_107738240 | 3.40 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr7_-_137028498 | 3.38 |
ENST00000393083.2
|
PTN
|
pleiotrophin |
| chr8_-_17533838 | 3.37 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr12_-_6233828 | 3.33 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr4_+_114214125 | 3.29 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr7_+_97361218 | 3.27 |
ENST00000319273.5
|
TAC1
|
tachykinin, precursor 1 |
| chr1_+_50569575 | 3.23 |
ENST00000371827.1
|
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_+_69985792 | 3.06 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_183774240 | 3.05 |
ENST00000360851.3
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr2_-_50201327 | 3.00 |
ENST00000412315.1
|
NRXN1
|
neurexin 1 |
| chr8_-_33457453 | 2.98 |
ENST00000523956.1
ENST00000256261.4 |
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chr12_-_89746173 | 2.89 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr1_-_68698222 | 2.86 |
ENST00000370976.3
ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr7_+_97361388 | 2.83 |
ENST00000350485.4
ENST00000346867.4 |
TAC1
|
tachykinin, precursor 1 |
| chr19_-_7968427 | 2.83 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr10_-_50970322 | 2.80 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr2_-_50574856 | 2.79 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr2_+_68962014 | 2.73 |
ENST00000467265.1
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_-_74618964 | 2.71 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr19_+_50016411 | 2.65 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr9_+_12693336 | 2.61 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr3_+_69985734 | 2.45 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr12_-_10282742 | 2.37 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_+_100136811 | 2.35 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr2_+_90248739 | 2.35 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr1_-_45956800 | 2.34 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr3_-_33686743 | 2.33 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr1_-_152386732 | 2.33 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr3_+_173116225 | 2.31 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr17_-_77924627 | 2.28 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr9_+_5890802 | 2.26 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr2_-_74619152 | 2.21 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr10_-_50970382 | 2.12 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr4_+_72204755 | 2.10 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chrX_-_19988382 | 2.05 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr14_+_29236269 | 2.02 |
ENST00000313071.4
|
FOXG1
|
forkhead box G1 |
| chr11_-_128894053 | 2.01 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr10_+_18629628 | 2.01 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr12_+_26348429 | 2.00 |
ENST00000242729.2
|
SSPN
|
sarcospan |
| chr9_+_82186872 | 1.98 |
ENST00000376544.3
ENST00000376520.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr19_-_53758094 | 1.96 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr7_-_14026063 | 1.88 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chrX_+_107288239 | 1.80 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_-_32908792 | 1.79 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr6_+_153552455 | 1.78 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr8_+_105235572 | 1.78 |
ENST00000523362.1
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr8_-_18541603 | 1.78 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_150738261 | 1.77 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr5_-_20575959 | 1.75 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr2_-_145277569 | 1.74 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr3_-_114790179 | 1.71 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr9_+_470288 | 1.70 |
ENST00000382303.1
|
KANK1
|
KN motif and ankyrin repeat domains 1 |
| chr11_-_108093329 | 1.69 |
ENST00000278612.8
|
NPAT
|
nuclear protein, ataxia-telangiectasia locus |
| chr12_+_12938541 | 1.69 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr12_+_7014126 | 1.67 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr12_+_7014064 | 1.65 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr12_-_118797475 | 1.63 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr2_+_103035102 | 1.63 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr10_+_74451883 | 1.62 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr7_+_49813255 | 1.62 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr7_-_22233442 | 1.62 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr13_-_88323218 | 1.61 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr13_-_95131923 | 1.58 |
ENST00000377028.5
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr19_-_51522955 | 1.56 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr9_-_215744 | 1.56 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr19_+_54641444 | 1.51 |
ENST00000221232.5
ENST00000358389.3 |
CNOT3
|
CCR4-NOT transcription complex, subunit 3 |
| chr3_-_114477962 | 1.48 |
ENST00000471418.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr17_+_41363854 | 1.48 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr2_+_68961934 | 1.48 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chrX_+_107288197 | 1.43 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_-_14026123 | 1.43 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr12_-_118796910 | 1.42 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr4_+_88754069 | 1.42 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr19_+_34287751 | 1.41 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr3_-_114477787 | 1.41 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr4_-_41884620 | 1.40 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr18_+_32556892 | 1.39 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr18_-_500692 | 1.39 |
ENST00000400256.3
|
COLEC12
|
collectin sub-family member 12 |
| chr2_+_68961905 | 1.39 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr2_-_111334678 | 1.38 |
ENST00000329516.3
ENST00000330331.5 ENST00000446930.1 |
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr17_+_47448102 | 1.38 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr5_+_66300446 | 1.38 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr20_+_18488137 | 1.37 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr12_+_7013897 | 1.37 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr1_-_151762943 | 1.36 |
ENST00000368825.3
ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH
|
tudor and KH domain containing |
| chr9_-_21351377 | 1.35 |
ENST00000380210.1
|
IFNA6
|
interferon, alpha 6 |
| chr8_+_119294456 | 1.31 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr4_-_74486347 | 1.31 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr12_-_10022735 | 1.29 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr4_+_3388057 | 1.29 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr3_+_69928256 | 1.28 |
ENST00000394355.2
|
MITF
|
microphthalmia-associated transcription factor |
| chr18_-_53253323 | 1.27 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chr7_-_14029515 | 1.26 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr4_+_41614909 | 1.26 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_-_145278475 | 1.26 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_+_69321074 | 1.25 |
ENST00000380751.5
ENST00000380750.3 ENST00000503931.1 ENST00000506542.1 |
SERF1B
|
small EDRK-rich factor 1B (centromeric) |
| chr8_+_9953061 | 1.23 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr8_+_101170563 | 1.23 |
ENST00000520508.1
ENST00000388798.2 |
SPAG1
|
sperm associated antigen 1 |
| chr7_+_138943265 | 1.22 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr7_-_151217001 | 1.19 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr18_-_53253112 | 1.19 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr7_-_14029283 | 1.18 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr8_+_117950422 | 1.18 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr8_+_9953214 | 1.14 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr14_-_82000140 | 1.14 |
ENST00000555824.1
ENST00000557372.1 ENST00000336735.4 |
SEL1L
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr7_-_83278322 | 1.13 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr12_+_81110684 | 1.10 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr1_+_160160283 | 1.09 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr2_+_171034646 | 1.08 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
| chr7_-_14028488 | 1.07 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr6_+_72922590 | 1.06 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_166930131 | 1.03 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr14_-_98444386 | 1.01 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr2_+_90211643 | 0.99 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr4_-_170897045 | 0.98 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr11_-_102576537 | 0.96 |
ENST00000260229.4
|
MMP27
|
matrix metallopeptidase 27 |
| chr17_+_72427477 | 0.95 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_-_28113965 | 0.95 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr14_-_92413727 | 0.94 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr1_+_226013047 | 0.93 |
ENST00000366837.4
|
EPHX1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr6_+_72922505 | 0.93 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr10_+_119301928 | 0.91 |
ENST00000553456.3
|
EMX2
|
empty spiracles homeobox 2 |
| chr4_+_88754113 | 0.90 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr2_-_208031943 | 0.90 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_+_26402465 | 0.89 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr18_+_46065393 | 0.89 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr2_+_210444142 | 0.88 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr10_+_13628933 | 0.88 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr12_-_23737534 | 0.87 |
ENST00000396007.2
|
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr8_-_87755878 | 0.86 |
ENST00000320005.5
|
CNGB3
|
cyclic nucleotide gated channel beta 3 |
| chr6_-_91296602 | 0.86 |
ENST00000369325.3
ENST00000369327.3 |
MAP3K7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr8_+_28748765 | 0.86 |
ENST00000355231.5
|
HMBOX1
|
homeobox containing 1 |
| chr10_+_102222798 | 0.84 |
ENST00000343737.5
|
WNT8B
|
wingless-type MMTV integration site family, member 8B |
| chr12_-_58135903 | 0.82 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr20_+_2276639 | 0.82 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr6_-_22297730 | 0.81 |
ENST00000306482.1
|
PRL
|
prolactin |
| chr6_+_26402517 | 0.79 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr10_-_71169031 | 0.79 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr19_-_51523275 | 0.78 |
ENST00000309958.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chr6_-_138539627 | 0.77 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr6_-_136788001 | 0.76 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr7_-_104909435 | 0.76 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr6_-_100912785 | 0.75 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr5_-_159827033 | 0.75 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr2_+_90273679 | 0.75 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr16_+_89334512 | 0.74 |
ENST00000602042.1
|
AC137932.1
|
AC137932.1 |
| chr12_-_51422017 | 0.74 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr1_-_101360331 | 0.73 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr12_+_18414446 | 0.73 |
ENST00000433979.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr7_-_22234381 | 0.73 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr8_-_72274467 | 0.72 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr13_-_99630233 | 0.72 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr2_-_30144432 | 0.72 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr4_-_74486217 | 0.71 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_+_59783941 | 0.71 |
ENST00000506884.1
ENST00000504876.2 |
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr12_-_10605929 | 0.70 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr14_+_74034310 | 0.69 |
ENST00000538782.1
|
ACOT2
|
acyl-CoA thioesterase 2 |
| chr20_-_21494654 | 0.69 |
ENST00000377142.4
|
NKX2-2
|
NK2 homeobox 2 |
| chrX_+_105937068 | 0.67 |
ENST00000324342.3
|
RNF128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr19_+_45458503 | 0.65 |
ENST00000337392.5
ENST00000591304.1 |
CLPTM1
|
cleft lip and palate associated transmembrane protein 1 |
| chr5_+_125758813 | 0.64 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 25.9 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 2.3 | 23.2 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 2.3 | 6.9 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 2.0 | 6.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 1.2 | 2.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 1.1 | 3.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.0 | 3.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.9 | 6.3 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.8 | 65.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.8 | 8.4 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.8 | 14.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.8 | 17.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.7 | 3.0 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.7 | 5.8 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.7 | 2.9 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.7 | 2.0 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.6 | 4.9 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.6 | 1.8 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.6 | 1.7 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.5 | 2.6 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.5 | 9.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.4 | 6.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.4 | 8.9 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.4 | 6.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.4 | 3.7 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 4.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.3 | 3.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 1.7 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 2.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 0.9 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 0.9 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.3 | 2.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.7 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 0.8 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.3 | 1.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 2.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 0.8 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.2 | 1.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.2 | 0.7 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 0.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.2 | 1.6 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.2 | 3.8 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.2 | 0.6 | GO:2000007 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm development(GO:0048389) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) pattern specification involved in mesonephros development(GO:0061227) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in kidney development(GO:0072098) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter urothelium development(GO:0072190) ureter epithelial cell differentiation(GO:0072192) ureter morphogenesis(GO:0072197) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.2 | 0.6 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.2 | 0.6 | GO:0042109 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.5 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.2 | 0.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.2 | 0.5 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.2 | 0.7 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 3.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.2 | 0.6 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.2 | 0.8 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.7 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.9 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 1.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 5.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 6.8 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 1.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 2.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 2.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.9 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.1 | 0.9 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.2 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.1 | 0.3 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 1.8 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 2.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 4.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.3 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 1.4 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.8 | GO:2001259 | positive regulation of cation channel activity(GO:2001259) |
| 0.1 | 0.7 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 1.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 2.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 3.1 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.1 | 0.3 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 9.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 2.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 2.3 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 1.1 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.1 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.9 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 1.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 1.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 5.4 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.3 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 1.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 4.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.5 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 11.5 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 3.8 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 0.8 | GO:0001823 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.0 | 0.8 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 1.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.6 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.6 | GO:0033081 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 | 0.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.7 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 1.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 2.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 4.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.3 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 3.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.3 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 3.0 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 1.3 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.4 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.5 | GO:0002763 | positive regulation of myeloid leukocyte differentiation(GO:0002763) |
| 0.0 | 1.8 | GO:0009408 | response to heat(GO:0009408) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.9 | 14.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.7 | 4.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.6 | 3.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 4.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 4.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 6.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 0.9 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.3 | 4.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 1.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 3.6 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 1.8 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.2 | 2.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 1.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 1.7 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.2 | 1.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 5.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 4.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 2.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 1.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 2.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 36.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 8.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 9.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 6.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 2.0 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 21.1 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 16.2 | GO:0045121 | membrane raft(GO:0045121) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 3.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.8 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 59.4 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 5.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.9 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.8 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 3.3 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.6 | 6.3 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.2 | 68.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 1.0 | 3.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.0 | 2.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.0 | 4.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.9 | 3.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.8 | 7.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.6 | 3.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.6 | 7.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 2.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 14.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 2.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 5.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 9.0 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.3 | 1.2 | GO:0015265 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.3 | 0.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 0.8 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.3 | 1.6 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.3 | 2.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 0.6 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.2 | 1.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.2 | 2.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.5 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 8.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 2.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 4.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 0.5 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 6.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 0.8 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.2 | 3.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 5.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 6.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.7 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.7 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.1 | 4.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 4.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 3.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.9 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 3.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 2.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 4.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.7 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 1.4 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 24.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 6.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.9 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 3.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.3 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 3.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 8.8 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.1 | 1.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 16.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 4.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 4.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.6 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 3.1 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.3 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 7.7 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 13.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 6.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 3.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 7.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 6.0 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 10.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 5.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 1.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 4.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 3.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 2.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 2.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 4.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 4.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 11.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 2.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 6.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 5.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |