Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
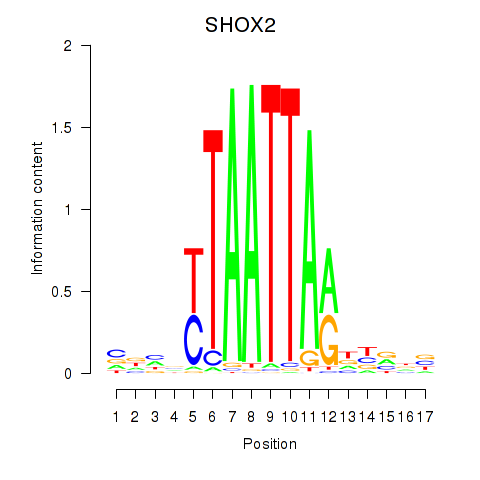
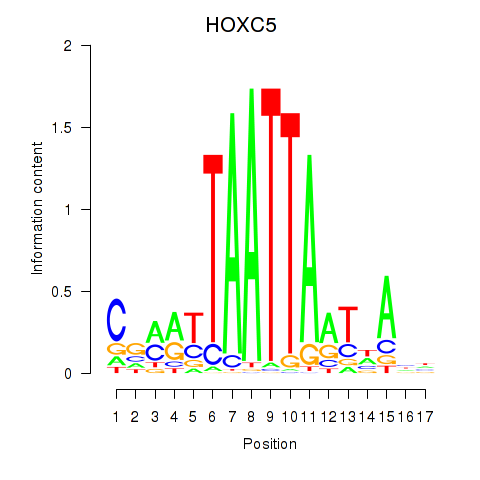
Results for SHOX2_HOXC5
Z-value: 0.24
Transcription factors associated with SHOX2_HOXC5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX2
|
ENSG00000168779.15 | short stature homeobox 2 |
|
HOXC5
|
ENSG00000172789.3 | homeobox C5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SHOX2 | hg19_v2_chr3_-_157824292_157824334 | -0.40 | 9.7e-10 | Click! |
| HOXC5 | hg19_v2_chr12_+_54426637_54426637 | -0.07 | 2.8e-01 | Click! |
Activity profile of SHOX2_HOXC5 motif
Sorted Z-values of SHOX2_HOXC5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_89292422 | 31.06 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr6_-_32557610 | 30.36 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr2_+_90108504 | 29.71 |
ENST00000390271.2
|
IGKV6D-41
|
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr2_+_90077680 | 29.35 |
ENST00000390270.2
|
IGKV3D-20
|
immunoglobulin kappa variable 3D-20 |
| chr1_-_153518270 | 24.44 |
ENST00000354332.4
ENST00000368716.4 |
S100A4
|
S100 calcium binding protein A4 |
| chr2_-_89385283 | 23.93 |
ENST00000390252.2
|
IGKV3-15
|
immunoglobulin kappa variable 3-15 |
| chr2_-_89340242 | 23.52 |
ENST00000480492.1
|
IGKV1-12
|
immunoglobulin kappa variable 1-12 |
| chr2_+_90198535 | 23.50 |
ENST00000390276.2
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr2_-_89247338 | 22.63 |
ENST00000496168.1
|
IGKV1-5
|
immunoglobulin kappa variable 1-5 |
| chr11_+_35201826 | 22.53 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr15_-_20193370 | 22.42 |
ENST00000558565.2
|
IGHV3OR15-7
|
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr2_-_89327228 | 20.67 |
ENST00000483158.1
|
IGKV3-11
|
immunoglobulin kappa variable 3-11 |
| chr7_-_87856280 | 20.57 |
ENST00000490437.1
ENST00000431660.1 |
SRI
|
sorcin |
| chr7_+_115862858 | 20.43 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr2_-_89513402 | 20.23 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr2_+_87565634 | 20.20 |
ENST00000421835.2
|
IGKV3OR2-268
|
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr2_+_90153696 | 20.09 |
ENST00000417279.2
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr2_+_90139056 | 19.94 |
ENST00000492446.1
|
IGKV1D-16
|
immunoglobulin kappa variable 1D-16 |
| chr2_-_90538397 | 19.31 |
ENST00000443397.3
|
RP11-685N3.1
|
Uncharacterized protein |
| chr2_+_90273679 | 19.03 |
ENST00000423080.2
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr7_-_87856303 | 18.56 |
ENST00000394641.3
|
SRI
|
sorcin |
| chr11_+_5710919 | 18.47 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr11_-_104817919 | 18.26 |
ENST00000533252.1
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr2_+_90248739 | 17.62 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr6_+_26365443 | 16.92 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chr11_-_104972158 | 16.83 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr17_+_48823975 | 16.66 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr12_-_10022735 | 16.62 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr7_+_141438118 | 16.45 |
ENST00000265304.6
ENST00000498107.1 ENST00000467681.1 ENST00000465582.1 ENST00000463093.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr3_+_37035289 | 16.28 |
ENST00000455445.2
ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1
|
mutL homolog 1 |
| chr14_-_106552755 | 16.22 |
ENST00000390600.2
|
IGHV3-9
|
immunoglobulin heavy variable 3-9 |
| chr2_+_68961934 | 15.87 |
ENST00000409202.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr14_-_107049312 | 15.87 |
ENST00000390627.2
|
IGHV3-53
|
immunoglobulin heavy variable 3-53 |
| chr7_-_139763521 | 15.82 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr2_+_68961905 | 15.66 |
ENST00000295381.3
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr4_+_113568207 | 15.05 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_-_89357179 | 14.40 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr4_-_103749179 | 14.38 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_33913708 | 14.11 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr2_-_17981462 | 13.82 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr16_+_12059091 | 13.32 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr6_+_31802364 | 13.19 |
ENST00000375640.3
ENST00000375641.2 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chr4_-_103749205 | 13.14 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_89521942 | 12.68 |
ENST00000482769.1
|
IGKV2-28
|
immunoglobulin kappa variable 2-28 |
| chr5_-_10761206 | 12.64 |
ENST00000432074.2
ENST00000230895.6 |
DAP
|
death-associated protein |
| chr2_+_219110149 | 12.61 |
ENST00000456575.1
|
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chrX_-_100872911 | 12.31 |
ENST00000361910.4
ENST00000539247.1 ENST00000538627.1 |
ARMCX6
|
armadillo repeat containing, X-linked 6 |
| chr11_-_104827425 | 12.29 |
ENST00000393150.3
|
CASP4
|
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_42584847 | 12.22 |
ENST00000372883.3
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_-_89417335 | 12.15 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr7_-_102184083 | 12.05 |
ENST00000379357.5
|
POLR2J3
|
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr6_+_32407619 | 11.82 |
ENST00000395388.2
ENST00000374982.5 |
HLA-DRA
|
major histocompatibility complex, class II, DR alpha |
| chr14_+_52456327 | 11.82 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr2_-_89568263 | 11.65 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr2_+_89952792 | 11.63 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr2_+_90211643 | 11.53 |
ENST00000390277.2
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr6_+_139456226 | 11.39 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr5_-_176433350 | 11.11 |
ENST00000377227.4
ENST00000377219.2 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr2_-_89160770 | 11.01 |
ENST00000390240.2
|
IGKJ3
|
immunoglobulin kappa joining 3 |
| chr3_+_138340049 | 10.88 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr11_-_104905840 | 10.87 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_+_220267429 | 10.72 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr19_-_14945933 | 10.70 |
ENST00000322301.3
|
OR7A5
|
olfactory receptor, family 7, subfamily A, member 5 |
| chr14_-_106668095 | 10.58 |
ENST00000390606.2
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr6_+_31802685 | 10.56 |
ENST00000375639.2
ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chr1_-_160681593 | 10.30 |
ENST00000368045.3
ENST00000368046.3 |
CD48
|
CD48 molecule |
| chr15_+_64680003 | 10.17 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr10_+_122610687 | 10.15 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr2_-_176046391 | 10.02 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr20_-_33735070 | 9.87 |
ENST00000374491.3
ENST00000542871.1 ENST00000374492.3 |
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr8_-_101718991 | 9.81 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr22_+_21996549 | 9.80 |
ENST00000248958.4
|
SDF2L1
|
stromal cell-derived factor 2-like 1 |
| chr12_-_15114603 | 9.70 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_-_158345462 | 9.67 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr17_+_66511540 | 9.58 |
ENST00000588188.2
|
PRKAR1A
|
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_-_197115818 | 9.51 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr6_+_26104104 | 9.47 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr6_-_41909561 | 9.39 |
ENST00000372991.4
|
CCND3
|
cyclin D3 |
| chr18_-_47018897 | 9.25 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr18_-_47018869 | 9.20 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr14_-_106926724 | 9.17 |
ENST00000434710.1
|
IGHV3-43
|
immunoglobulin heavy variable 3-43 |
| chr19_-_39826639 | 8.81 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr14_+_35591928 | 8.80 |
ENST00000605870.1
ENST00000557404.3 |
KIAA0391
|
KIAA0391 |
| chr7_+_120591170 | 8.65 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr15_+_58430567 | 8.61 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chr19_+_50016610 | 8.53 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_+_32077386 | 8.50 |
ENST00000354689.6
|
IGHV3OR16-9
|
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr21_-_15918618 | 8.43 |
ENST00000400564.1
ENST00000400566.1 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_+_113558612 | 8.35 |
ENST00000505034.1
ENST00000324052.6 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr8_-_90996837 | 8.31 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr1_-_24306835 | 8.18 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr3_-_141719195 | 8.05 |
ENST00000397991.4
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_+_12059050 | 8.01 |
ENST00000396495.3
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr17_+_7155819 | 7.98 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr3_-_141747950 | 7.97 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_-_23426337 | 7.94 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr2_-_238323007 | 7.92 |
ENST00000295550.4
|
COL6A3
|
collagen, type VI, alpha 3 |
| chr11_+_125462690 | 7.90 |
ENST00000392708.4
ENST00000529196.1 ENST00000531491.1 |
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr5_-_81574160 | 7.89 |
ENST00000510210.1
ENST00000512493.1 ENST00000507980.1 ENST00000511844.1 ENST00000510019.1 |
RPS23
|
ribosomal protein S23 |
| chr19_-_17516449 | 7.86 |
ENST00000252593.6
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr2_+_86333301 | 7.84 |
ENST00000254630.7
|
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr14_-_23426322 | 7.80 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr2_-_89597542 | 7.64 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr17_+_41150290 | 7.63 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr1_-_1709845 | 7.60 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr1_-_150738261 | 7.60 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr2_-_89278535 | 7.60 |
ENST00000390247.2
|
IGKV3-7
|
immunoglobulin kappa variable 3-7 (non-functional) |
| chr4_+_113558272 | 7.58 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr19_+_50016411 | 7.57 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_+_48823896 | 7.56 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr12_-_102455846 | 7.52 |
ENST00000545679.1
|
CCDC53
|
coiled-coil domain containing 53 |
| chr19_+_23945768 | 7.50 |
ENST00000486528.1
ENST00000496398.1 |
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr19_+_48248779 | 7.50 |
ENST00000246802.5
|
GLTSCR2
|
glioma tumor suppressor candidate region gene 2 |
| chr8_-_109260897 | 7.40 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr14_-_23426270 | 7.39 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr4_-_47465666 | 7.37 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr3_-_30936153 | 7.34 |
ENST00000454381.3
ENST00000282538.5 |
GADL1
|
glutamate decarboxylase-like 1 |
| chr6_-_109702885 | 7.32 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr1_-_1710229 | 7.26 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr16_+_1359511 | 7.22 |
ENST00000397514.3
ENST00000397515.2 ENST00000567383.1 ENST00000403747.2 ENST00000566587.1 |
UBE2I
|
ubiquitin-conjugating enzyme E2I |
| chr13_+_37581115 | 7.22 |
ENST00000481013.1
|
EXOSC8
|
exosome component 8 |
| chr6_+_26440700 | 7.22 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr2_+_187371440 | 7.21 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr14_-_106573756 | 7.20 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr12_+_75874460 | 7.09 |
ENST00000266659.3
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr18_-_47018769 | 7.06 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chrX_-_122756660 | 7.06 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr2_+_89901292 | 7.02 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr4_-_83812248 | 6.99 |
ENST00000514326.1
ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr3_+_138340067 | 6.93 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr16_-_58585513 | 6.90 |
ENST00000245138.4
ENST00000567285.1 |
CNOT1
|
CCR4-NOT transcription complex, subunit 1 |
| chr18_-_51751132 | 6.86 |
ENST00000256429.3
|
MBD2
|
methyl-CpG binding domain protein 2 |
| chr3_+_148709128 | 6.84 |
ENST00000345003.4
ENST00000296048.6 ENST00000483267.1 |
GYG1
|
glycogenin 1 |
| chr1_-_205091115 | 6.84 |
ENST00000264515.6
ENST00000367164.1 |
RBBP5
|
retinoblastoma binding protein 5 |
| chr12_-_31479045 | 6.84 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr6_+_63921399 | 6.83 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr5_+_177631523 | 6.78 |
ENST00000506339.1
ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr3_-_16524357 | 6.78 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_+_158801095 | 6.78 |
ENST00000368141.4
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr10_-_58120996 | 6.78 |
ENST00000361148.6
ENST00000395405.1 ENST00000373944.3 |
ZWINT
|
ZW10 interacting kinetochore protein |
| chr1_-_145826450 | 6.77 |
ENST00000462900.2
|
GPR89A
|
G protein-coupled receptor 89A |
| chr14_-_100841930 | 6.75 |
ENST00000555031.1
ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr4_-_103749313 | 6.68 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_94451574 | 6.65 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr5_+_177631497 | 6.65 |
ENST00000358344.3
|
HNRNPAB
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr17_+_41150479 | 6.63 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chr1_+_158979792 | 6.61 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr14_-_20801427 | 6.61 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr5_-_142780280 | 6.54 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr21_-_35284635 | 6.53 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr1_-_183559693 | 6.52 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_-_193075180 | 6.48 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr21_+_10862622 | 6.45 |
ENST00000302092.5
ENST00000559480.1 |
IGHV1OR21-1
|
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chrX_-_16887963 | 6.43 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr3_+_37034823 | 6.39 |
ENST00000231790.2
ENST00000456676.2 |
MLH1
|
mutL homolog 1 |
| chr19_-_13227463 | 6.39 |
ENST00000437766.1
ENST00000221504.8 |
TRMT1
|
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr1_+_158979680 | 6.34 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr17_-_30228678 | 6.33 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr7_+_142498725 | 6.27 |
ENST00000466254.1
|
TRBC2
|
T cell receptor beta constant 2 |
| chr1_+_158979686 | 6.26 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr6_-_31508304 | 6.26 |
ENST00000376177.2
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr12_-_9913489 | 6.25 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr2_+_102413726 | 6.22 |
ENST00000350878.4
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr11_-_14521379 | 6.21 |
ENST00000249923.3
ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1
|
coatomer protein complex, subunit beta 1 |
| chr5_+_110074685 | 6.19 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr16_-_66864806 | 6.18 |
ENST00000566336.1
ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr17_+_8191815 | 6.15 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr12_+_56435637 | 6.11 |
ENST00000356464.5
ENST00000552361.1 |
RPS26
|
ribosomal protein S26 |
| chr6_+_34204642 | 6.10 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr8_-_101719159 | 6.09 |
ENST00000520868.1
ENST00000522658.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr2_+_102456277 | 6.09 |
ENST00000421882.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_+_104337515 | 6.05 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr2_-_158300556 | 6.04 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr4_-_103748880 | 5.97 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_83812402 | 5.97 |
ENST00000395310.2
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr7_-_64023441 | 5.95 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr1_-_35450897 | 5.86 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr14_+_35761580 | 5.83 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr7_-_22862406 | 5.78 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr19_-_10491234 | 5.77 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr11_-_71823715 | 5.76 |
ENST00000545944.1
ENST00000502597.2 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr5_+_96038476 | 5.71 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr20_+_1099233 | 5.67 |
ENST00000246015.4
ENST00000335877.6 ENST00000438768.2 |
PSMF1
|
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr9_-_128246769 | 5.66 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chrX_-_109590174 | 5.65 |
ENST00000372054.1
|
GNG5P2
|
guanine nucleotide binding protein (G protein), gamma 5 pseudogene 2 |
| chr1_-_212965104 | 5.62 |
ENST00000422588.2
ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1
|
NSL1, MIS12 kinetochore complex component |
| chr9_+_108463234 | 5.61 |
ENST00000374688.1
|
TMEM38B
|
transmembrane protein 38B |
| chr1_-_39339777 | 5.60 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr4_-_74853897 | 5.57 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr2_+_90259593 | 5.57 |
ENST00000471857.1
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr4_-_103749105 | 5.55 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr19_-_10227503 | 5.54 |
ENST00000593054.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr2_+_207024306 | 5.51 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_+_95523621 | 5.47 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr5_-_149829244 | 5.46 |
ENST00000312037.5
|
RPS14
|
ribosomal protein S14 |
| chr1_+_53480598 | 5.41 |
ENST00000430330.2
ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2
|
sterol carrier protein 2 |
| chr2_-_89310012 | 5.41 |
ENST00000493819.1
|
IGKV1-9
|
immunoglobulin kappa variable 1-9 |
| chr3_-_64009658 | 5.39 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr8_-_90996459 | 5.38 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr15_+_80351910 | 5.37 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX2_HOXC5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.0 | 26.9 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 8.4 | 42.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 6.9 | 27.7 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 5.6 | 39.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 3.6 | 7.2 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 3.6 | 10.7 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 3.4 | 30.4 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 3.3 | 9.9 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 3.2 | 13.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 2.8 | 22.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 2.7 | 13.6 | GO:0071918 | pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 2.7 | 8.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 2.6 | 15.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 2.6 | 7.9 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 2.5 | 14.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 2.4 | 9.7 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 2.4 | 14.4 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 2.3 | 16.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.3 | 6.8 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 2.2 | 6.7 | GO:0061010 | gall bladder development(GO:0061010) |
| 2.2 | 332.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 2.1 | 6.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 2.0 | 12.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 1.9 | 11.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 1.8 | 7.2 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 1.8 | 5.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 1.8 | 15.9 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.7 | 5.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 1.7 | 17.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 1.6 | 16.5 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 1.6 | 9.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 1.6 | 4.7 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 1.5 | 4.6 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 1.5 | 6.2 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 1.5 | 6.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 1.4 | 4.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 1.4 | 7.1 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.4 | 6.8 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 1.3 | 12.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 1.3 | 3.9 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 1.3 | 5.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 1.3 | 5.2 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 1.3 | 5.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.3 | 5.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 1.3 | 8.8 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 1.2 | 5.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.2 | 6.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 1.2 | 3.6 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 1.2 | 19.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.2 | 18.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 1.2 | 3.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 1.2 | 4.6 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 1.2 | 5.8 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 1.1 | 3.3 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 1.1 | 4.4 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.1 | 5.4 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 1.1 | 13.0 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 1.1 | 3.2 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 1.1 | 10.5 | GO:0042262 | DNA protection(GO:0042262) |
| 1.0 | 161.8 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 1.0 | 3.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.0 | 3.1 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 1.0 | 45.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 1.0 | 4.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 1.0 | 6.9 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 1.0 | 6.8 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.9 | 5.7 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.9 | 5.5 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.9 | 18.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.9 | 30.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.9 | 2.7 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.9 | 2.7 | GO:0039516 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.9 | 5.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.9 | 22.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.9 | 4.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.9 | 2.6 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.9 | 3.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.8 | 4.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.8 | 7.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.8 | 3.3 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.8 | 4.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.8 | 6.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.8 | 6.5 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.8 | 3.8 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.8 | 3.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.7 | 3.0 | GO:1904504 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.7 | 2.2 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.7 | 2.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.7 | 2.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.7 | 5.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.7 | 5.7 | GO:0001878 | response to yeast(GO:0001878) |
| 0.7 | 2.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.7 | 9.6 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.7 | 0.7 | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining(GO:2001033) |
| 0.7 | 3.3 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.6 | 4.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.6 | 14.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.6 | 2.5 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.6 | 11.9 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.6 | 17.5 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.6 | 2.5 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.6 | 3.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.6 | 1.8 | GO:0060920 | atrioventricular node development(GO:0003162) cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.6 | 2.4 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.6 | 7.2 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.6 | 3.6 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.6 | 5.9 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.6 | 1.8 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.6 | 10.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 4.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.6 | 4.0 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.6 | 4.0 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.6 | 7.3 | GO:0008652 | cellular amino acid biosynthetic process(GO:0008652) |
| 0.6 | 5.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 1.6 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.5 | 13.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 4.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 8.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.5 | 1.6 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.5 | 5.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 2.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.5 | 6.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.5 | 7.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.5 | 10.8 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.5 | 4.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.5 | 5.0 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.5 | 1.5 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.5 | 3.9 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.5 | 10.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.5 | 6.1 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.5 | 4.7 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.5 | 7.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.5 | 6.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 11.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.5 | 2.3 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.5 | 0.9 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.5 | 3.7 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.5 | 3.7 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.5 | 0.9 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.5 | 1.4 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.4 | 11.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.4 | 7.1 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.4 | 3.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.4 | 19.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.4 | 14.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.4 | 25.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.4 | 2.2 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.4 | 1.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.4 | 3.0 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.4 | 3.3 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.4 | 56.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.4 | 0.8 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.4 | 1.2 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.4 | 1.2 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.4 | 1.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.4 | 1.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.4 | 1.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.4 | 1.5 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.3 | 17.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.3 | 2.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 4.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.3 | 33.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.3 | 2.0 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.3 | 9.4 | GO:0002479 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) |
| 0.3 | 27.7 | GO:1904667 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051439) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 2.3 | GO:0009615 | response to virus(GO:0009615) |
| 0.3 | 1.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.3 | 10.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 2.3 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 1.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.3 | 3.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.3 | 3.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.3 | 2.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.3 | 6.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.3 | 2.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.3 | 8.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 8.0 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.3 | 2.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 2.4 | GO:0015816 | glycine transport(GO:0015816) |
| 0.3 | 2.6 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.3 | 1.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.3 | 1.7 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 2.6 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.3 | 2.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.3 | 6.1 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.3 | 1.9 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.3 | 4.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 7.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 18.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.3 | 19.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 1.8 | GO:0044092 | negative regulation of molecular function(GO:0044092) |
| 0.3 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 3.0 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.2 | 3.4 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 0.7 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 9.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 2.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 2.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.2 | 0.9 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.2 | 2.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.2 | 2.1 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.2 | 2.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 5.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.2 | 1.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 0.7 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 9.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.2 | 2.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.2 | 1.0 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 5.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 1.2 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) |
| 0.2 | 2.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.2 | 2.2 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.2 | 4.0 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 | 3.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.2 | 0.6 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.2 | 0.6 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.2 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 6.8 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.2 | 0.8 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.2 | 1.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 4.1 | GO:0006294 | nucleotide-excision repair, preincision complex assembly(GO:0006294) |
| 0.2 | 5.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 0.4 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.2 | 3.0 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 2.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 13.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.2 | 1.4 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.2 | 2.4 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 5.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.2 | 7.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 2.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 2.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 2.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 1.5 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 4.2 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 0.8 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 | 1.9 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 6.5 | GO:0043069 | negative regulation of apoptotic process(GO:0043066) negative regulation of programmed cell death(GO:0043069) |
| 0.2 | 3.6 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.2 | 2.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 23.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.2 | 0.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.2 | 0.8 | GO:0030070 | insulin processing(GO:0030070) |
| 0.2 | 1.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 1.0 | GO:0071569 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 2.6 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 3.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 1.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 3.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.1 | 3.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 2.2 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 1.6 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 6.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.0 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.1 | 0.4 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.1 | 4.9 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.1 | 1.0 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 2.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.8 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.1 | 2.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 3.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 6.1 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.6 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.1 | 2.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 5.0 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.1 | 2.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.1 | 6.6 | GO:0035722 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.1 | 1.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.3 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 4.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.5 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 1.1 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.1 | 1.0 | GO:0060850 | regulation of transcription involved in cell fate commitment(GO:0060850) |
| 0.1 | 1.7 | GO:0010659 | cardiac muscle cell apoptotic process(GO:0010659) |
| 0.1 | 1.0 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.1 | 4.4 | GO:0061053 | somite development(GO:0061053) |
| 0.1 | 6.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 13.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 2.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 1.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.6 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.1 | 1.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.1 | 4.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 3.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 2.1 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.1 | 2.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 1.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.7 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.1 | 0.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.8 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.1 | 1.7 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 4.3 | GO:0051452 | intracellular pH reduction(GO:0051452) |
| 0.1 | 3.9 | GO:1904894 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.3 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 4.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.5 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 6.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 3.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 2.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 2.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 2.1 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 1.9 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.2 | GO:0061356 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) negative regulation of Wnt protein secretion(GO:0061358) |
| 0.1 | 26.8 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.1 | 8.9 | GO:0050851 | antigen receptor-mediated signaling pathway(GO:0050851) |
| 0.1 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 1.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 4.9 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.5 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 2.2 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.7 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.5 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.5 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 2.0 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.4 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.8 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.6 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.7 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.5 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.7 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.6 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.2 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 55.6 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 9.0 | 26.9 | GO:0005715 | late recombination nodule(GO:0005715) |
| 5.6 | 39.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 4.2 | 12.6 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 3.5 | 13.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 3.2 | 22.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 3.0 | 15.2 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 2.6 | 23.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 2.3 | 7.0 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 2.3 | 9.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 2.2 | 8.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 2.1 | 42.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 2.0 | 14.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.9 | 13.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.8 | 1.8 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.5 | 4.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.4 | 9.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.3 | 6.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 1.3 | 66.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.3 | 4.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 1.3 | 14.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.3 | 17.8 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 1.1 | 17.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 1.1 | 5.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.1 | 2.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.1 | 7.6 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 1.0 | 8.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.0 | 4.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 1.0 | 11.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.0 | 7.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 1.0 | 5.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.0 | 4.8 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 1.0 | 6.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.9 | 8.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.9 | 4.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.9 | 11.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.8 | 15.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.8 | 5.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.8 | 9.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.8 | 6.5 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.8 | 4.0 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.7 | 3.0 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.7 | 16.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.7 | 17.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.7 | 6.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.7 | 6.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.7 | 3.4 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.7 | 7.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.7 | 2.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.7 | 7.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.7 | 3.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.6 | 1.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.6 | 3.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.6 | 21.3 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.6 | 6.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.6 | 4.9 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.6 | 1.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.6 | 15.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.6 | 10.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.6 | 6.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.6 | 5.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 2.7 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.5 | 3.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.5 | 1.5 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.5 | 9.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 3.0 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.5 | 3.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.5 | 7.4 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.5 | 9.8 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.5 | 1.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.5 | 4.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.5 | 6.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.4 | 31.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.4 | 80.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.4 | 5.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.4 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 3.0 | GO:0043203 | axon hillock(GO:0043203) |
| 0.4 | 5.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 5.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.4 | 12.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 2.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 41.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.4 | 3.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 2.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 1.5 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.4 | 1.5 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 1.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.4 | 1.4 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.3 | 4.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.3 | 4.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.3 | 18.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 3.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.3 | 3.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.3 | 1.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.3 | 16.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.3 | 7.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 20.0 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.3 | 5.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 2.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.3 | 4.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 3.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.3 | 5.7 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.3 | 1.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.3 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.3 | 7.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.2 | 2.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.9 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.2 | 39.3 | GO:0101002 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.2 | 0.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 2.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.6 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 5.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.2 | 1.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 9.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 3.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 8.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 8.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.1 | GO:0098574 | cytoplasmic side of late endosome membrane(GO:0098560) cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.2 | 1.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 3.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 3.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.2 | 12.9 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.2 | 2.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.2 | 5.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 3.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 10.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 0.9 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 3.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 6.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 1.8 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 2.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 7.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 0.3 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.1 | 2.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 2.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 21.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 3.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 4.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.1 | 1.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 3.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0036019 | endolysosome(GO:0036019) |
| 0.1 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.8 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 2.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 10.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 0.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 8.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) nBAF complex(GO:0071565) |
| 0.1 | 4.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 4.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 34.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 133.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 4.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 7.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 19.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 34.2 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.1 | 2.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.3 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 4.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 5.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 4.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.6 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 9.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 1.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 8.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.1 | GO:0098552 | side of membrane(GO:0098552) |
| 0.0 | 1.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 1.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 58.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 4.0 | 16.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 3.6 | 10.7 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 3.4 | 13.6 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 3.4 | 26.9 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 2.8 | 11.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 2.3 | 9.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 2.2 | 6.7 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 2.2 | 10.9 | GO:0070728 | leucine binding(GO:0070728) |
| 2.2 | 4.4 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 2.2 | 503.8 | GO:0003823 | antigen binding(GO:0003823) |
| 1.9 | 5.8 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 1.9 | 9.5 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 1.9 | 13.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 1.8 | 7.3 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 1.8 | 7.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 1.8 | 5.4 | GO:0070538 | oleic acid binding(GO:0070538) |
| 1.8 | 5.3 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 1.7 | 6.8 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 1.7 | 11.9 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 1.7 | 13.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 1.6 | 9.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 1.6 | 4.7 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 1.5 | 12.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.4 | 4.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 1.4 | 7.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 1.4 | 4.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 1.4 | 21.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 1.4 | 6.8 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 1.3 | 5.3 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 1.3 | 6.5 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 1.2 | 5.0 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.2 | 24.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.2 | 9.7 | GO:0046790 | virion binding(GO:0046790) |
| 1.2 | 4.7 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 1.1 | 6.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.1 | 5.6 | GO:0070404 | NADH binding(GO:0070404) |
| 1.1 | 8.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 1.1 | 7.7 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 1.1 | 3.3 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 1.1 | 49.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 1.0 | 1.0 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 1.0 | 4.0 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 1.0 | 3.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.0 | 9.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.0 | 37.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 1.0 | 3.8 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 1.0 | 5.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.9 | 15.9 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 4.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.9 | 3.7 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.9 | 2.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.9 | 2.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.9 | 5.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.9 | 3.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.9 | 6.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.8 | 5.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.8 | 9.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.8 | 2.5 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.8 | 10.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.8 | 7.9 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 2.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.7 | 21.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.7 | 4.8 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.7 | 2.7 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.7 | 6.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.7 | 3.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.6 | 5.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.6 | 4.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.6 | 2.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.6 | 10.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.6 | 3.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.6 | 3.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.6 | 4.1 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.6 | 6.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.6 | 5.8 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.6 | 12.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.6 | 22.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.6 | 17.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.6 | 7.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.6 | 8.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.6 | 13.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.5 | 4.9 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.5 | 9.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.5 | 2.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 1.6 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.5 | 2.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.5 | 3.0 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 1.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.5 | 9.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 5.8 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.5 | 5.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.5 | 0.9 | GO:0046977 | peptide antigen-transporting ATPase activity(GO:0015433) TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 0.5 | 2.8 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.5 | 3.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 1.4 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.4 | 4.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 18.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.4 | 1.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.4 | 12.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.4 | 12.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.4 | 3.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.4 | 11.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 6.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 1.2 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.4 | 14.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 2.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 3.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 6.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 8.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.4 | 2.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.4 | 6.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.4 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 1.4 | GO:0047718 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.3 | 5.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 5.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.3 | 1.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 9.8 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 3.6 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.3 | 69.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.3 | 1.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.3 | 3.2 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.3 | 5.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 2.4 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 19.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 1.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.3 | 1.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.3 | 2.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 3.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 2.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.3 | 2.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.3 | 1.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.3 | 2.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 4.9 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.3 | 5.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.3 | 5.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 16.5 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.3 | 2.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 14.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.3 | 6.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 1.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 5.7 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.3 | 1.0 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.3 | 2.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 5.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.2 | 3.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 2.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 5.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 5.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 6.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 0.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 5.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 1.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.2 | 3.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.8 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.2 | 4.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 4.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 3.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 1.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.6 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.2 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 10.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 5.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 1.9 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 18.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 7.9 | GO:0033558 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.2 | 2.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 4.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 4.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 4.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.2 | 0.5 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 5.5 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.2 | 2.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 1.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 2.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 26.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.2 | 8.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 0.5 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 1.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 4.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0004549 | tRNA-specific ribonuclease activity(GO:0004549) |
| 0.1 | 2.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 21.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 4.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 3.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 6.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.5 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.1 | 1.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 3.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 8.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 0.6 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 8.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.6 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) ferrous iron transmembrane transporter activity(GO:0015093) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 2.7 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 1.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 2.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 2.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 22.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 1.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 5.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.1 | 0.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 5.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 6.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 3.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.8 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 5.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 20.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 16.4 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 2.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 2.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 62.9 | PID BARD1 PATHWAY | BARD1 signaling events |
| 1.2 | 32.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.9 | 43.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.8 | 22.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.6 | 5.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.6 | 8.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 2.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.5 | 21.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.4 | 10.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.4 | 14.3 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 9.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.3 | 4.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 20.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.2 | 10.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 10.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.2 | 9.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 5.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.2 | 30.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.2 | 8.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 5.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 15.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.2 | 5.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 6.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.2 | 9.3 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.2 | 8.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 6.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 11.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 4.5 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 4.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 5.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 7.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 3.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 3.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.1 | 2.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 2.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 8.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 33.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 1.4 | 67.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 1.3 | 47.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 1.2 | 23.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.9 | 12.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.8 | 9.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.8 | 7.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.8 | 10.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.7 | 11.7 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.7 | 6.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.6 | 11.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.6 | 13.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.6 | 61.7 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.6 | 6.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.6 | 19.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.5 | 10.7 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.5 | 33.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.4 | 2.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.4 | 41.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.4 | 39.8 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.4 | 5.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 11.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 2.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 2.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.3 | 5.8 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 20.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 5.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 4.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 6.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 9.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 5.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 4.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 6.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 7.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 3.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 4.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 14.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 7.3 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 2.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 7.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 16.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 1.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 9.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.2 | 2.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 2.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.2 | 1.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 4.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 5.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.2 | 1.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.2 | 2.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 3.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 9.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 10.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 5.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 15.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 8.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 8.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 21.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.5 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 3.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 14.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 4.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 4.1 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 2.7 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.4 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.9 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 3.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.2 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |