Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
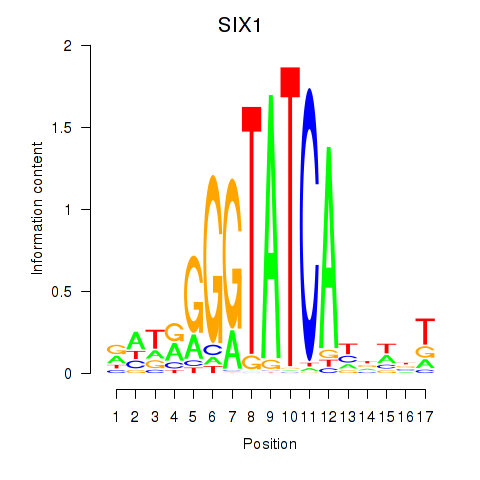
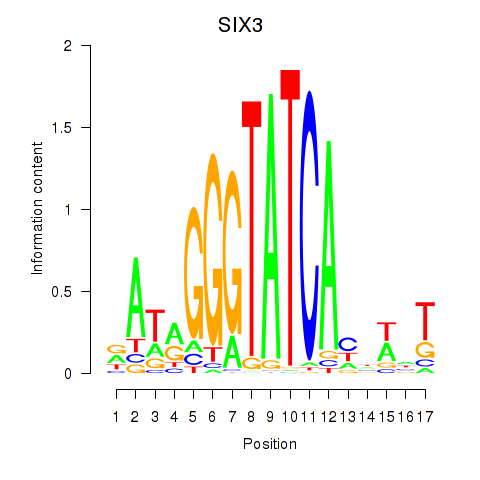
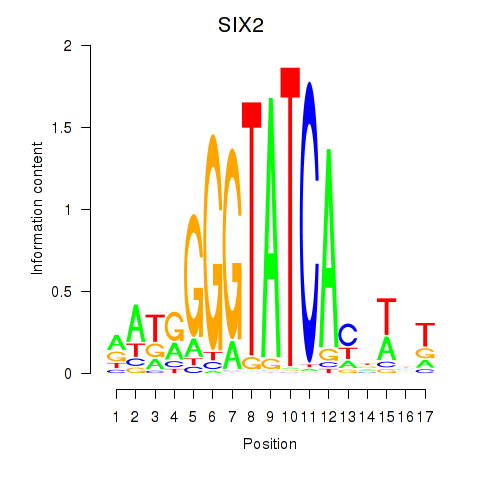
Results for SIX1_SIX3_SIX2
Z-value: 0.57
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.72 | 4.2e-36 | Click! |
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | 0.67 | 7.2e-30 | Click! |
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.37 | 2.1e-08 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_139025875 | 15.19 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr3_+_52448539 | 10.99 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr11_+_70244510 | 8.80 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr16_-_28550348 | 8.78 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr21_+_27011584 | 8.44 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr3_+_42850959 | 8.22 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr1_-_13390765 | 7.99 |
ENST00000357367.2
|
PRAMEF8
|
PRAME family member 8 |
| chr17_+_1665345 | 7.93 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr3_+_38323785 | 7.87 |
ENST00000466887.1
ENST00000448498.1 |
SLC22A14
|
solute carrier family 22, member 14 |
| chr1_-_183387723 | 7.66 |
ENST00000287713.6
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr16_-_28550320 | 7.53 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr21_+_27011899 | 7.47 |
ENST00000425221.2
|
JAM2
|
junctional adhesion molecule 2 |
| chr14_+_75761099 | 7.41 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr17_-_41466555 | 7.40 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr11_-_58499434 | 7.09 |
ENST00000344743.3
ENST00000278400.3 |
GLYAT
|
glycine-N-acyltransferase |
| chr1_-_158656488 | 6.96 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chrX_+_56259316 | 6.93 |
ENST00000468660.1
|
KLF8
|
Kruppel-like factor 8 |
| chr1_+_169337172 | 6.91 |
ENST00000367807.3
ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr1_-_149982624 | 6.75 |
ENST00000417191.1
ENST00000369135.4 |
OTUD7B
|
OTU domain containing 7B |
| chr11_-_111794446 | 6.52 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr12_-_6798616 | 6.00 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr6_-_167797887 | 5.98 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr11_+_71900572 | 5.69 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
| chr11_-_32452357 | 5.63 |
ENST00000379079.2
ENST00000530998.1 |
WT1
|
Wilms tumor 1 |
| chr3_+_111718036 | 5.59 |
ENST00000455401.2
|
TAGLN3
|
transgelin 3 |
| chr1_+_152975488 | 5.56 |
ENST00000542696.1
|
SPRR3
|
small proline-rich protein 3 |
| chr1_+_241695424 | 5.52 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr12_-_58220078 | 5.35 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_207119738 | 5.03 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr1_+_160709029 | 4.93 |
ENST00000444090.2
ENST00000441662.2 |
SLAMF7
|
SLAM family member 7 |
| chrX_+_13587712 | 4.82 |
ENST00000361306.1
ENST00000380602.3 |
EGFL6
|
EGF-like-domain, multiple 6 |
| chr7_-_44198874 | 4.81 |
ENST00000395796.3
ENST00000345378.2 |
GCK
|
glucokinase (hexokinase 4) |
| chr1_+_160709055 | 4.81 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr1_+_241695670 | 4.81 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr3_+_111718173 | 4.75 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr1_-_13117736 | 4.58 |
ENST00000376192.5
ENST00000376182.1 |
PRAMEF6
|
PRAME family member 6 |
| chr1_+_156698708 | 4.51 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr3_+_111717600 | 4.37 |
ENST00000273368.4
|
TAGLN3
|
transgelin 3 |
| chr1_+_22964073 | 4.33 |
ENST00000402322.1
|
C1QA
|
complement component 1, q subcomponent, A chain |
| chr1_+_182419261 | 4.33 |
ENST00000294854.8
ENST00000542961.1 |
RGSL1
|
regulator of G-protein signaling like 1 |
| chr11_+_73661364 | 4.32 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr1_-_54665664 | 4.23 |
ENST00000542737.1
ENST00000537208.1 |
CYB5RL
|
cytochrome b5 reductase-like |
| chr6_+_131894284 | 4.21 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr1_-_13115578 | 4.19 |
ENST00000414205.2
|
PRAMEF6
|
PRAME family member 6 |
| chr5_-_132073210 | 4.19 |
ENST00000378735.1
ENST00000378746.4 |
KIF3A
|
kinesin family member 3A |
| chr15_-_90892669 | 4.07 |
ENST00000412799.2
|
GABARAPL3
|
GABA(A) receptors associated protein like 3, pseudogene |
| chr11_-_76155618 | 4.06 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr1_+_12916941 | 3.96 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr1_+_196946680 | 3.94 |
ENST00000256785.4
|
CFHR5
|
complement factor H-related 5 |
| chr12_-_118796910 | 3.92 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr1_-_109618566 | 3.88 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr1_-_85462623 | 3.85 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr6_+_88757507 | 3.83 |
ENST00000237201.1
|
SPACA1
|
sperm acrosome associated 1 |
| chr6_-_52710893 | 3.79 |
ENST00000284562.2
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr11_-_128712362 | 3.79 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr15_+_40331456 | 3.76 |
ENST00000504245.1
ENST00000560341.1 |
SRP14-AS1
|
SRP14 antisense RNA1 (head to head) |
| chr11_-_2906979 | 3.73 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr18_-_51750948 | 3.71 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr3_+_184529929 | 3.71 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr13_-_46679185 | 3.70 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr1_-_12946025 | 3.64 |
ENST00000235349.5
|
PRAMEF4
|
PRAME family member 4 |
| chr2_+_105953972 | 3.59 |
ENST00000410049.1
|
C2orf49
|
chromosome 2 open reading frame 49 |
| chr1_+_13359819 | 3.59 |
ENST00000376168.1
|
PRAMEF5
|
PRAME family member 5 |
| chr9_+_6716478 | 3.58 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr2_+_234826016 | 3.54 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr11_-_76155700 | 3.50 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr10_+_7745303 | 3.47 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr1_-_12958101 | 3.45 |
ENST00000235347.4
|
PRAMEF10
|
PRAME family member 10 |
| chr1_+_35225339 | 3.43 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chr13_-_46679144 | 3.42 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr12_+_57857475 | 3.40 |
ENST00000528467.1
|
GLI1
|
GLI family zinc finger 1 |
| chr7_+_100551239 | 3.29 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr1_+_158901329 | 3.29 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr19_-_50316517 | 3.27 |
ENST00000313777.4
ENST00000445575.2 |
FUZ
|
fuzzy planar cell polarity protein |
| chr9_+_132427883 | 3.22 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chrX_+_114524275 | 3.21 |
ENST00000371921.1
ENST00000451986.2 ENST00000371920.3 |
LUZP4
|
leucine zipper protein 4 |
| chr19_+_45445491 | 3.20 |
ENST00000592954.1
ENST00000419266.2 ENST00000589057.1 |
APOC4
APOC4-APOC2
|
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr1_+_15256230 | 3.18 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr11_-_35547151 | 3.17 |
ENST00000378878.3
ENST00000529303.1 ENST00000278360.3 |
PAMR1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr11_+_64323428 | 3.17 |
ENST00000377581.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr11_+_12132117 | 3.16 |
ENST00000256194.4
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr17_-_6915646 | 3.16 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr15_+_80733570 | 3.15 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_196946664 | 3.14 |
ENST00000367414.5
|
CFHR5
|
complement factor H-related 5 |
| chr11_+_64323098 | 3.10 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr10_+_104613980 | 3.06 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr11_-_71791518 | 3.05 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr10_+_7745232 | 3.05 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr22_+_19710468 | 3.02 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr1_+_160709076 | 2.99 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr11_+_64323156 | 2.96 |
ENST00000377585.3
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr3_+_184529948 | 2.94 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr14_+_95047725 | 2.93 |
ENST00000554760.1
ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr5_+_125758813 | 2.86 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_+_16330723 | 2.86 |
ENST00000329454.2
|
C1orf64
|
chromosome 1 open reading frame 64 |
| chr2_-_73053126 | 2.82 |
ENST00000272427.6
ENST00000410104.1 |
EXOC6B
|
exocyst complex component 6B |
| chr1_+_150245177 | 2.77 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr19_+_2867325 | 2.76 |
ENST00000307635.2
ENST00000586426.1 |
ZNF556
|
zinc finger protein 556 |
| chr1_+_12976450 | 2.75 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr12_+_110011571 | 2.75 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr19_-_3600549 | 2.72 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chrX_+_138612889 | 2.72 |
ENST00000218099.2
ENST00000394090.2 |
F9
|
coagulation factor IX |
| chr10_+_48255253 | 2.68 |
ENST00000357718.4
ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8
AL591684.1
|
annexin A8 Protein LOC100996760 |
| chr16_+_1306060 | 2.65 |
ENST00000397534.2
|
TPSD1
|
tryptase delta 1 |
| chr22_-_44258360 | 2.62 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr10_+_47746929 | 2.56 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr3_-_10452359 | 2.54 |
ENST00000452124.1
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr1_+_13736907 | 2.53 |
ENST00000316412.5
|
PRAMEF20
|
PRAME family member 20 |
| chr19_+_45445524 | 2.52 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C-IV |
| chr17_-_59668550 | 2.51 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr10_-_47173994 | 2.45 |
ENST00000414655.2
ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1
LINC00842
|
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr1_+_26737292 | 2.45 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr12_+_81471816 | 2.44 |
ENST00000261206.3
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3 |
| chrX_+_101906294 | 2.43 |
ENST00000361600.5
ENST00000415986.1 ENST00000444152.1 ENST00000537097.1 |
GPRASP1
|
G protein-coupled receptor associated sorting protein 1 |
| chr13_+_28712614 | 2.38 |
ENST00000380958.3
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr13_+_53226963 | 2.38 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr10_+_5005445 | 2.38 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr20_+_36946029 | 2.37 |
ENST00000417318.1
|
BPI
|
bactericidal/permeability-increasing protein |
| chr19_+_57999079 | 2.37 |
ENST00000426954.2
ENST00000354197.4 ENST00000523882.1 ENST00000520540.1 ENST00000519310.1 ENST00000442920.2 ENST00000523312.1 ENST00000424930.2 |
ZNF419
|
zinc finger protein 419 |
| chr19_+_57999101 | 2.35 |
ENST00000347466.6
ENST00000523138.1 ENST00000415379.2 ENST00000521754.1 ENST00000221735.7 ENST00000518999.1 ENST00000521137.1 |
ZNF419
|
zinc finger protein 419 |
| chr3_+_149191723 | 2.35 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr3_+_35722487 | 2.34 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr19_+_55476620 | 2.31 |
ENST00000543010.1
ENST00000391721.4 ENST00000339757.7 |
NLRP2
|
NLR family, pyrin domain containing 2 |
| chr19_+_42055879 | 2.31 |
ENST00000407170.2
ENST00000601116.1 ENST00000595395.1 |
CEACAM21
AC006129.2
|
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chr16_-_15463926 | 2.31 |
ENST00000432570.2
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr1_+_12851545 | 2.30 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr1_-_12891264 | 2.27 |
ENST00000535591.1
ENST00000437584.1 |
PRAMEF11
|
PRAME family member 11 |
| chr14_-_95942173 | 2.27 |
ENST00000334258.5
ENST00000557275.1 ENST00000553340.1 |
SYNE3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr11_-_49230144 | 2.26 |
ENST00000343844.4
|
FOLH1
|
folate hydrolase (prostate-specific membrane antigen) 1 |
| chr8_+_18248786 | 2.26 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr16_+_27214802 | 2.25 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr11_+_117947782 | 2.23 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr8_-_19540086 | 2.21 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr19_-_11039261 | 2.13 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr17_+_4675175 | 2.12 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr2_+_48844973 | 2.12 |
ENST00000403751.3
|
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr20_+_34700333 | 2.12 |
ENST00000441639.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chrX_+_101470280 | 2.10 |
ENST00000395088.2
ENST00000330252.5 ENST00000333110.5 |
NXF2
TCP11X1
|
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chrX_+_8432871 | 2.09 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr8_+_18248755 | 2.09 |
ENST00000286479.3
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr16_-_68014732 | 2.09 |
ENST00000268793.4
|
DPEP3
|
dipeptidase 3 |
| chr16_+_56666563 | 2.08 |
ENST00000570233.1
|
MT1M
|
metallothionein 1M |
| chr8_-_18666360 | 2.08 |
ENST00000286485.8
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr6_+_73076432 | 2.08 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr9_-_79307096 | 2.07 |
ENST00000376717.2
ENST00000223609.6 ENST00000443509.2 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr15_+_62853562 | 2.07 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr11_-_59633951 | 2.04 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr7_-_6866401 | 2.03 |
ENST00000316731.8
|
CCZ1B
|
CCZ1 vacuolar protein trafficking and biogenesis associated homolog B (S. cerevisiae) |
| chr1_+_26737253 | 2.00 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr16_+_56642041 | 2.00 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr7_-_14880892 | 1.99 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr11_-_44972476 | 1.99 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr9_-_34662651 | 1.98 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr19_+_41594377 | 1.98 |
ENST00000330436.3
|
CYP2A13
|
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr12_+_69186125 | 1.96 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr4_+_155484155 | 1.96 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr11_+_125365110 | 1.96 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr7_-_73133959 | 1.95 |
ENST00000395155.3
ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A
|
syntaxin 1A (brain) |
| chr9_-_6645628 | 1.93 |
ENST00000321612.6
|
GLDC
|
glycine dehydrogenase (decarboxylating) |
| chr7_+_75677354 | 1.93 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr2_-_232395169 | 1.91 |
ENST00000305141.4
|
NMUR1
|
neuromedin U receptor 1 |
| chr5_+_125758865 | 1.90 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr1_+_120049826 | 1.88 |
ENST00000369413.3
ENST00000235547.6 ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr11_-_44972390 | 1.86 |
ENST00000395648.3
ENST00000531928.2 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr18_+_61575200 | 1.84 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr3_-_9834375 | 1.82 |
ENST00000343450.2
ENST00000301964.2 |
TADA3
|
transcriptional adaptor 3 |
| chr2_-_114514181 | 1.82 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr19_-_15344243 | 1.82 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr1_-_13007420 | 1.81 |
ENST00000376189.1
|
PRAMEF6
|
PRAME family member 6 |
| chr11_-_44972418 | 1.81 |
ENST00000525680.1
ENST00000528290.1 ENST00000530035.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr2_+_48844937 | 1.80 |
ENST00000448460.1
ENST00000437125.1 ENST00000430487.2 |
GTF2A1L
|
general transcription factor IIA, 1-like |
| chr1_-_154150651 | 1.79 |
ENST00000302206.5
|
TPM3
|
tropomyosin 3 |
| chr2_-_152589670 | 1.78 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr10_+_51549498 | 1.77 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr1_-_159046617 | 1.75 |
ENST00000368130.4
|
AIM2
|
absent in melanoma 2 |
| chr11_-_71791435 | 1.75 |
ENST00000351960.6
ENST00000541719.1 ENST00000535111.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr10_+_5005598 | 1.74 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr12_-_89919965 | 1.74 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr5_+_81575281 | 1.72 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr8_-_19540266 | 1.72 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr11_+_117947724 | 1.67 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr12_-_89920030 | 1.67 |
ENST00000413530.1
ENST00000547474.1 |
GALNT4
POC1B-GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr2_+_211342432 | 1.67 |
ENST00000430249.2
|
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr6_-_136847099 | 1.67 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr8_+_99129513 | 1.65 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr19_-_48389651 | 1.64 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr18_-_11148587 | 1.63 |
ENST00000302079.6
ENST00000580640.1 ENST00000503781.3 |
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr21_+_35736302 | 1.62 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr11_+_114168085 | 1.61 |
ENST00000541754.1
|
NNMT
|
nicotinamide N-methyltransferase |
| chr17_+_43213004 | 1.61 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr19_+_15852203 | 1.61 |
ENST00000305892.1
|
OR10H3
|
olfactory receptor, family 10, subfamily H, member 3 |
| chr7_+_29519486 | 1.60 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr16_+_451826 | 1.59 |
ENST00000219481.5
ENST00000397710.1 ENST00000424398.2 |
DECR2
|
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr12_-_58135903 | 1.59 |
ENST00000257897.3
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr3_-_46506358 | 1.58 |
ENST00000417439.1
ENST00000431944.1 |
LTF
|
lactotransferrin |
| chr19_+_46800289 | 1.58 |
ENST00000377670.4
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit |
| chr19_-_11039188 | 1.57 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr1_+_196743912 | 1.56 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr9_-_21077939 | 1.55 |
ENST00000380232.2
|
IFNB1
|
interferon, beta 1, fibroblast |
| chr11_-_118134997 | 1.55 |
ENST00000278937.2
|
MPZL2
|
myelin protein zero-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 1.9 | 5.7 | GO:0060974 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 1.8 | 7.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.6 | 4.8 | GO:1902846 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 1.6 | 7.9 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 1.4 | 5.6 | GO:0072302 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 1.3 | 1.3 | GO:0030168 | platelet activation(GO:0030168) |
| 1.3 | 16.3 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 1.1 | 3.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 1.0 | 9.2 | GO:0015747 | urate transport(GO:0015747) |
| 1.0 | 5.0 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 1.0 | 3.9 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.9 | 2.7 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.9 | 2.7 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.9 | 2.7 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.9 | 4.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.9 | 7.7 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.9 | 3.4 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.8 | 6.8 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.8 | 4.2 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.8 | 3.3 | GO:2000314 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.8 | 5.7 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.8 | 8.8 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.8 | 3.8 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.7 | 5.0 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 3.5 | GO:0050955 | thermoception(GO:0050955) |
| 0.7 | 2.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.7 | 4.8 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.7 | 2.7 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.6 | 2.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.5 | 3.7 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.5 | 5.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.5 | 1.6 | GO:2000308 | iron assimilation(GO:0033212) iron assimilation by chelation and transport(GO:0033214) positive regulation of bone mineralization involved in bone maturation(GO:1900159) negative regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000308) |
| 0.5 | 4.7 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.5 | 3.1 | GO:0034201 | arginine biosynthetic process(GO:0006526) response to oleic acid(GO:0034201) |
| 0.5 | 7.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.5 | 1.0 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.5 | 2.0 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.5 | 2.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.5 | 4.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.4 | 1.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 2.4 | GO:0052055 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.4 | 0.8 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.4 | 1.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.4 | 1.9 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 2.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 1.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.4 | 6.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.4 | 1.1 | GO:1990535 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) neuron projection maintenance(GO:1990535) |
| 0.3 | 2.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.3 | 2.7 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.3 | 2.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.3 | 1.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.3 | 2.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.3 | 2.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.3 | 9.6 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.3 | 1.6 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.3 | 1.1 | GO:2000570 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.3 | 1.1 | GO:2001153 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.3 | 1.6 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.3 | 1.8 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.2 | 1.2 | GO:1900005 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.2 | 2.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.2 | 4.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 2.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 0.9 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.2 | 1.8 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 4.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 2.4 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.2 | 1.1 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.2 | 1.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.2 | 0.8 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.2 | 8.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 2.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 2.4 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 1.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 2.0 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.0 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.2 | 1.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 1.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.2 | 2.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 2.8 | GO:0042711 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.2 | 2.8 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.2 | 0.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 1.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.6 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.6 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.2 | 1.1 | GO:0007320 | insemination(GO:0007320) |
| 0.2 | 0.5 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 0.9 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.2 | 0.6 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 1.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 2.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 1.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 1.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 3.0 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 2.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.1 | 1.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.9 | GO:2001268 | positive regulation of keratinocyte migration(GO:0051549) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.1 | 4.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 5.0 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 5.0 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 4.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 2.1 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 4.3 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.1 | 1.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.6 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 2.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.1 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 1.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.6 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 6.7 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 4.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 1.5 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 | 2.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 5.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 8.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 5.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 2.3 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 3.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 4.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.6 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 1.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.1 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 1.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 9.0 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 0.6 | GO:0010885 | regulation of cholesterol storage(GO:0010885) positive regulation of cholesterol storage(GO:0010886) |
| 0.1 | 3.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.7 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 12.4 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.1 | 2.4 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 2.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 1.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.7 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.1 | 0.8 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 | 2.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) primary lung bud formation(GO:0060431) lung induction(GO:0060492) |
| 0.1 | 2.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 4.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 35.4 | GO:0045596 | negative regulation of cell differentiation(GO:0045596) |
| 0.0 | 0.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 1.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 1.7 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 1.5 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 3.5 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.9 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 7.5 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.7 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.5 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 1.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.9 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 6.2 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 1.8 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 1.0 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.2 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.4 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.9 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 1.6 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 1.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 9.8 | GO:0007417 | central nervous system development(GO:0007417) |
| 0.0 | 1.4 | GO:0007267 | cell-cell signaling(GO:0007267) |
| 0.0 | 1.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0061673 | cortical microtubule(GO:0055028) mitotic spindle astral microtubule(GO:0061673) |
| 1.2 | 3.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 1.1 | 5.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 1.0 | 4.2 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.0 | 7.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.0 | 2.9 | GO:0036029 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.9 | 4.3 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.8 | 2.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 7.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.6 | 1.9 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.6 | 6.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.5 | 3.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.5 | 2.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 1.8 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 6.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 2.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.4 | 6.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.4 | 2.5 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.2 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.2 | 1.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 1.9 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.2 | 3.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.2 | 1.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 5.7 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 2.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 3.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.2 | 8.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 5.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 1.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 4.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 1.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 4.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 14.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 15.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 3.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 10.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 5.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 15.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 3.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 6.6 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.1 | 4.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 4.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 12.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 10.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 4.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.3 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 2.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 5.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 1.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 2.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 3.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 6.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 3.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.6 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 11.2 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.7 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 1.4 | 5.7 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 1.3 | 3.9 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 1.2 | 5.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 1.1 | 5.6 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.1 | 4.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 1.1 | 5.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 1.0 | 3.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.9 | 9.2 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.9 | 2.7 | GO:0004960 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.9 | 4.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.8 | 3.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.8 | 2.3 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.8 | 3.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.7 | 10.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.7 | 6.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.6 | 3.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.6 | 8.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.6 | 3.8 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.6 | 5.0 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.6 | 1.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 4.8 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.6 | 2.4 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.6 | 2.3 | GO:1904493 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.5 | 1.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.5 | 8.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.5 | 0.9 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.4 | 7.9 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.4 | 4.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.4 | 1.5 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 1.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 4.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 5.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 4.8 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.3 | 1.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 1.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.3 | 2.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 1.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.3 | 1.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 1.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.3 | 2.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 7.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 2.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 2.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 3.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 1.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.6 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.2 | 7.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 3.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 1.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 3.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 2.0 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 0.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.2 | 1.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.2 | 5.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 0.8 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.2 | 2.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.2 | 0.9 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.2 | 2.0 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 2.4 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.2 | 0.9 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.2 | 15.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 2.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.6 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 3.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 3.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 2.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 3.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 2.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 1.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 20.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.1 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 8.9 | GO:0016410 | N-acyltransferase activity(GO:0016410) |
| 0.1 | 1.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 3.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 1.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.9 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 3.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 8.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 1.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 5.7 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.1 | 0.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 3.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 1.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 2.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.5 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.0 | 6.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 23.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 3.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 1.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 4.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 2.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 3.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 5.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.9 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 16.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 7.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 9.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 6.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 22.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 12.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.8 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 5.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 7.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 9.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 4.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.3 | 4.8 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 5.7 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.3 | 2.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 4.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 7.0 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 19.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 2.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 4.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 15.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 2.0 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.1 | 2.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.8 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 2.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 4.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 5.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 5.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.8 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 3.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 2.1 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 5.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 6.7 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.9 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.7 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |