Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
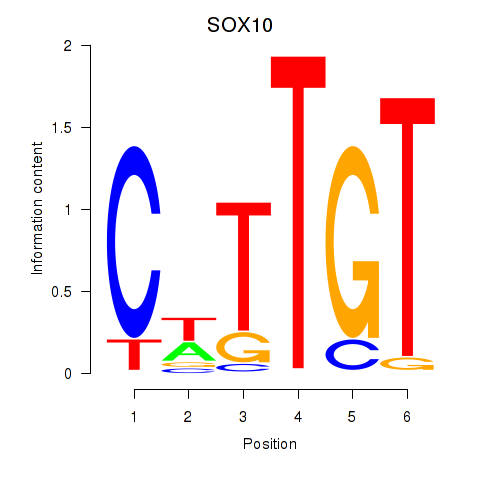
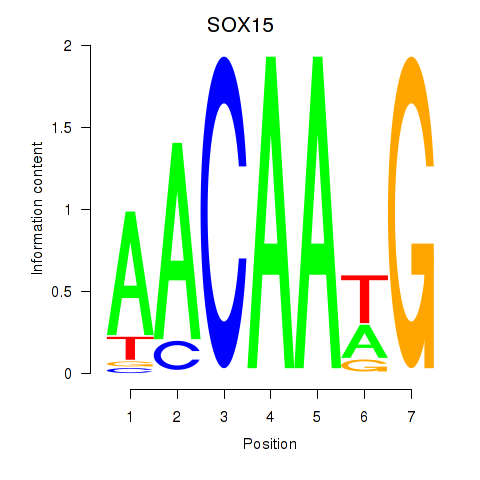
Results for SOX10_SOX15
Z-value: 0.46
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.39 | 3.2e-09 | Click! |
| SOX10 | hg19_v2_chr22_-_38380543_38380569 | -0.28 | 3.7e-05 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_80993010 | 16.90 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr13_-_41593425 | 16.55 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr15_+_57210961 | 14.34 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
| chr3_+_151986709 | 14.09 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr1_-_26233423 | 13.35 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr4_+_154387480 | 12.74 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chr2_+_10262857 | 11.83 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr17_+_36861735 | 11.74 |
ENST00000378137.5
ENST00000325718.7 |
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr20_+_35202909 | 10.88 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr2_+_232573222 | 10.60 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr10_+_11206925 | 10.32 |
ENST00000354440.2
ENST00000315874.4 ENST00000427450.1 |
CELF2
|
CUGBP, Elav-like family member 2 |
| chr2_-_10588630 | 9.93 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr5_-_175964366 | 9.88 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr7_+_23146271 | 9.49 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr4_-_109090106 | 9.39 |
ENST00000379951.2
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr2_+_232573208 | 9.03 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr15_+_52311398 | 8.77 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr2_+_120517174 | 8.60 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr1_-_32801825 | 8.38 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chrX_+_12993336 | 8.32 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr14_-_65409502 | 8.22 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr20_+_60718785 | 8.09 |
ENST00000421564.1
ENST00000450482.1 ENST00000331758.3 |
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr6_+_119215308 | 7.95 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr17_-_7297519 | 7.49 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr14_-_35344093 | 7.46 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chrX_+_12993202 | 7.42 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr11_+_125496619 | 7.39 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr17_-_7297833 | 7.14 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_-_200992827 | 7.02 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr5_+_159848807 | 6.99 |
ENST00000352433.5
|
PTTG1
|
pituitary tumor-transforming 1 |
| chr12_+_69979113 | 6.60 |
ENST00000299300.6
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr12_+_104359641 | 6.43 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr20_-_45984401 | 6.32 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_+_69979210 | 6.12 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr14_+_97263641 | 6.10 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr16_+_84801852 | 6.03 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr14_+_20923350 | 6.01 |
ENST00000555414.1
ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr17_+_75283973 | 5.98 |
ENST00000431235.2
ENST00000449803.2 |
SEPT9
|
septin 9 |
| chr12_+_69979446 | 5.81 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr13_+_111767650 | 5.80 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr4_-_83280767 | 5.76 |
ENST00000514671.1
|
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr14_-_65409438 | 5.71 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr14_-_71107921 | 5.70 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr3_-_18466026 | 5.59 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chrX_+_70503433 | 5.56 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr3_-_141747950 | 5.49 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_+_30689350 | 5.46 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr11_-_102323489 | 5.35 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr10_+_22605374 | 5.20 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr2_-_85625857 | 5.20 |
ENST00000453973.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr10_+_114710211 | 5.13 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_69004619 | 5.02 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr7_-_27219849 | 5.01 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chrX_-_135962876 | 5.00 |
ENST00000431446.3
ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX
|
RNA binding motif protein, X-linked |
| chrX_+_70503037 | 4.95 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr11_-_102323740 | 4.92 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr15_+_57210818 | 4.92 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chr4_-_109089573 | 4.88 |
ENST00000265165.1
|
LEF1
|
lymphoid enhancer-binding factor 1 |
| chr17_-_49124230 | 4.79 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr2_+_181845843 | 4.78 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr5_+_162930114 | 4.70 |
ENST00000280969.5
|
MAT2B
|
methionine adenosyltransferase II, beta |
| chr13_+_30002846 | 4.63 |
ENST00000542829.1
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr5_+_137514834 | 4.46 |
ENST00000508792.1
ENST00000504621.1 |
KIF20A
|
kinesin family member 20A |
| chr7_+_116660246 | 4.46 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr2_-_176046391 | 4.45 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr15_+_75074410 | 4.43 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr12_+_104359576 | 4.43 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr1_-_151431909 | 4.39 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr14_-_35099315 | 4.38 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr19_+_10765699 | 4.30 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr17_+_75447326 | 4.19 |
ENST00000591088.1
|
SEPT9
|
septin 9 |
| chr2_-_37899323 | 4.19 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr2_+_169312725 | 4.12 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr3_+_52719936 | 4.02 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr1_+_166808692 | 3.96 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr20_-_45985172 | 3.95 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_-_76953284 | 3.94 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr5_+_137514687 | 3.92 |
ENST00000394894.3
|
KIF20A
|
kinesin family member 20A |
| chr3_+_111260954 | 3.89 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr21_-_40720974 | 3.84 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr15_+_80364901 | 3.82 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr5_-_111093167 | 3.80 |
ENST00000446294.2
ENST00000419114.2 |
NREP
|
neuronal regeneration related protein |
| chr2_-_153573887 | 3.79 |
ENST00000493468.2
ENST00000545856.1 |
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr7_+_44788430 | 3.77 |
ENST00000457123.1
ENST00000309315.4 |
ZMIZ2
|
zinc finger, MIZ-type containing 2 |
| chr21_-_40720995 | 3.77 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chrX_+_80457442 | 3.76 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr2_+_114647504 | 3.72 |
ENST00000263238.2
|
ACTR3
|
ARP3 actin-related protein 3 homolog (yeast) |
| chr20_-_4804244 | 3.72 |
ENST00000379400.3
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr4_-_24586140 | 3.69 |
ENST00000336812.4
|
DHX15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr12_-_49582593 | 3.68 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr17_-_73389737 | 3.65 |
ENST00000392563.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr11_-_64013663 | 3.60 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr2_+_70056762 | 3.57 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr11_+_63706444 | 3.54 |
ENST00000377793.4
ENST00000456907.2 ENST00000539656.1 |
NAA40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr13_-_24007815 | 3.52 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr10_+_22605304 | 3.51 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr11_-_10830463 | 3.39 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr11_+_128563652 | 3.36 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr5_-_111092930 | 3.30 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr7_-_148581251 | 3.30 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr14_+_23791159 | 3.25 |
ENST00000557702.1
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr4_-_74088800 | 3.21 |
ENST00000509867.2
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr1_+_41174988 | 3.16 |
ENST00000372652.1
|
NFYC
|
nuclear transcription factor Y, gamma |
| chr17_+_27071002 | 3.13 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr14_+_52456193 | 3.13 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr9_+_131451480 | 3.11 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr5_+_43602750 | 3.06 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr7_+_111846643 | 3.03 |
ENST00000361822.3
|
ZNF277
|
zinc finger protein 277 |
| chr11_+_63754294 | 3.03 |
ENST00000543988.1
|
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr12_-_109027643 | 3.03 |
ENST00000388962.3
ENST00000550948.1 |
SELPLG
|
selectin P ligand |
| chr1_-_35658736 | 2.98 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr6_+_27114861 | 2.92 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr18_+_3448455 | 2.90 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr2_-_39348137 | 2.90 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr10_-_104953009 | 2.84 |
ENST00000470299.1
ENST00000343289.5 |
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr17_-_73781567 | 2.84 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr4_-_164534657 | 2.77 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr15_+_65823092 | 2.77 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr1_+_32479430 | 2.71 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr12_-_122985494 | 2.70 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr1_-_113247543 | 2.64 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr1_-_46152174 | 2.64 |
ENST00000290795.3
ENST00000355105.3 |
GPBP1L1
|
GC-rich promoter binding protein 1-like 1 |
| chr19_+_41257084 | 2.60 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr5_-_157002775 | 2.60 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr10_+_114709999 | 2.58 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr5_-_111092873 | 2.55 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr5_+_43603229 | 2.53 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_-_8751068 | 2.52 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr3_+_152879985 | 2.51 |
ENST00000323534.2
|
RAP2B
|
RAP2B, member of RAS oncogene family |
| chr4_-_76598544 | 2.51 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_+_107241783 | 2.48 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_79944336 | 2.42 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr1_-_151431647 | 2.40 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr1_-_114355083 | 2.36 |
ENST00000261441.5
|
RSBN1
|
round spermatid basic protein 1 |
| chr11_+_63753883 | 2.36 |
ENST00000538426.1
ENST00000543004.1 |
OTUB1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr16_+_67906919 | 2.35 |
ENST00000358933.5
|
EDC4
|
enhancer of mRNA decapping 4 |
| chr4_-_83351294 | 2.35 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr7_+_23145884 | 2.33 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr1_-_234614849 | 2.28 |
ENST00000040877.1
|
TARBP1
|
TAR (HIV-1) RNA binding protein 1 |
| chr17_-_76713100 | 2.24 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr1_-_151162606 | 2.21 |
ENST00000354473.4
ENST00000368892.4 |
VPS72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr17_-_7590745 | 2.20 |
ENST00000514944.1
ENST00000503591.1 ENST00000455263.2 ENST00000420246.2 ENST00000445888.2 ENST00000509690.1 ENST00000604348.1 ENST00000269305.4 |
TP53
|
tumor protein p53 |
| chr1_+_192544857 | 2.19 |
ENST00000367459.3
ENST00000469578.2 |
RGS1
|
regulator of G-protein signaling 1 |
| chr4_-_143227088 | 2.19 |
ENST00000511838.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr6_-_27114577 | 2.19 |
ENST00000356950.1
ENST00000396891.4 |
HIST1H2BK
|
histone cluster 1, H2bk |
| chr7_-_140178726 | 2.16 |
ENST00000480552.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr9_-_86595105 | 2.16 |
ENST00000457156.1
ENST00000360384.5 ENST00000376263.3 |
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr15_-_50647347 | 2.13 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr2_+_32288725 | 2.12 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr19_+_47105309 | 2.10 |
ENST00000599839.1
ENST00000596362.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr14_-_51027838 | 2.09 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr5_-_111093081 | 2.05 |
ENST00000453526.2
ENST00000509427.1 |
NREP
|
neuronal regeneration related protein |
| chr6_+_30689401 | 2.03 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr15_-_70388943 | 2.03 |
ENST00000559048.1
ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr5_+_147774275 | 2.02 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr4_+_41614720 | 2.01 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr10_+_99079008 | 2.01 |
ENST00000371021.3
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas |
| chr19_+_19303572 | 2.01 |
ENST00000407360.3
ENST00000540981.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr20_+_11898507 | 2.00 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr2_-_161350305 | 1.99 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_+_111260856 | 1.99 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr16_+_1359511 | 1.97 |
ENST00000397514.3
ENST00000397515.2 ENST00000567383.1 ENST00000403747.2 ENST00000566587.1 |
UBE2I
|
ubiquitin-conjugating enzyme E2I |
| chr12_-_122985067 | 1.96 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr14_-_22005018 | 1.95 |
ENST00000546363.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr7_-_27142290 | 1.93 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr17_-_73389854 | 1.92 |
ENST00000578961.1
ENST00000392564.1 ENST00000582582.1 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr2_-_47572105 | 1.88 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr17_-_53046058 | 1.84 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr17_-_9940058 | 1.83 |
ENST00000585266.1
|
GAS7
|
growth arrest-specific 7 |
| chrX_-_110655306 | 1.82 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr14_-_50583271 | 1.81 |
ENST00000395860.2
ENST00000395859.2 |
VCPKMT
|
valosin containing protein lysine (K) methyltransferase |
| chr2_-_153573965 | 1.79 |
ENST00000448428.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr12_-_90049878 | 1.77 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chrX_-_110655391 | 1.76 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr12_-_90049828 | 1.75 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr1_+_68150744 | 1.74 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr1_-_115259337 | 1.74 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr7_+_115850547 | 1.73 |
ENST00000358204.4
ENST00000455989.1 ENST00000537767.1 |
TES
|
testis derived transcript (3 LIM domains) |
| chr1_-_226926864 | 1.73 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr13_-_99959641 | 1.66 |
ENST00000376414.4
|
GPR183
|
G protein-coupled receptor 183 |
| chr12_-_42631529 | 1.65 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr1_+_24117627 | 1.64 |
ENST00000400061.1
|
LYPLA2
|
lysophospholipase II |
| chr21_-_16437126 | 1.63 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr17_+_67498538 | 1.61 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr12_-_71551868 | 1.60 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr7_-_76247617 | 1.60 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr6_-_111888474 | 1.59 |
ENST00000368735.1
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr17_-_56065484 | 1.56 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr14_-_91884150 | 1.53 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr7_+_50344289 | 1.52 |
ENST00000413698.1
ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr9_-_134955246 | 1.51 |
ENST00000357028.2
ENST00000474263.1 ENST00000292035.5 |
MED27
|
mediator complex subunit 27 |
| chr16_+_14980632 | 1.51 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr15_+_77224045 | 1.51 |
ENST00000320963.5
ENST00000394883.3 |
RCN2
|
reticulocalbin 2, EF-hand calcium binding domain |
| chr17_-_79881408 | 1.51 |
ENST00000392366.3
|
MAFG
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr2_-_161349909 | 1.47 |
ENST00000392753.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr11_+_35198118 | 1.46 |
ENST00000525211.1
ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44
|
CD44 molecule (Indian blood group) |
| chr12_-_14133053 | 1.44 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chr18_+_32558208 | 1.43 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_-_150924121 | 1.42 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr9_+_137218362 | 1.41 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.5 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 2.9 | 14.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 2.7 | 10.9 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 2.6 | 13.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 2.5 | 9.9 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 2.2 | 15.7 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 2.0 | 6.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 2.0 | 5.9 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 1.9 | 5.8 | GO:1901355 | response to rapamycin(GO:1901355) |
| 1.8 | 10.5 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 1.7 | 10.3 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.4 | 1.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 1.3 | 7.7 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 1.2 | 3.7 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 1.2 | 7.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 1.2 | 6.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.2 | 4.7 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.1 | 5.6 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 1.1 | 3.3 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 1.1 | 3.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 1.0 | 3.0 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.9 | 4.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.8 | 3.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.8 | 8.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) negative regulation of chromatin silencing(GO:0031936) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.8 | 5.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.7 | 2.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) oligodendrocyte apoptotic process(GO:0097252) |
| 0.7 | 3.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.7 | 4.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.7 | 2.7 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.6 | 10.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.6 | 4.4 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.6 | 16.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.6 | 10.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.6 | 2.9 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.6 | 3.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.6 | 1.7 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.5 | 1.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.5 | 2.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.5 | 1.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.5 | 1.6 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.5 | 6.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 11.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 1.5 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.5 | 3.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 1.4 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.5 | 8.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.5 | 10.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.5 | 1.8 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.5 | 8.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.4 | 2.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.4 | 5.8 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.4 | 2.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.4 | 1.2 | GO:0060926 | atrioventricular node development(GO:0003162) cardiac pacemaker cell differentiation(GO:0060920) cardiac pacemaker cell development(GO:0060926) |
| 0.4 | 1.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.4 | 3.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 2.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.3 | 5.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 1.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 | 3.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 4.2 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.3 | 2.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.3 | 1.7 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 5.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 1.7 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 | 0.9 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 0.8 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.3 | 3.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.3 | 2.5 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 10.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.3 | 1.6 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.3 | 7.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.3 | 3.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.3 | 1.3 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.2 | 1.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 3.6 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 5.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 3.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 3.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.5 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 3.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 | 2.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 3.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 17.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.2 | 2.0 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 4.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 5.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.2 | 6.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 7.0 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.2 | 1.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 | 6.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.5 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.3 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.1 | 3.8 | GO:0072662 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 4.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 11.8 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 0.8 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 16.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.4 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.9 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.5 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 4.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 2.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 15.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 7.5 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.1 | 2.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.9 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.8 | GO:0032276 | regulation of gonadotropin secretion(GO:0032276) negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 1.6 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.1 | 1.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 2.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 5.6 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 8.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.1 | 2.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.8 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.5 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.3 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 1.8 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 2.0 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.1 | 7.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.1 | 2.0 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.7 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.1 | 2.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 4.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 1.7 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 1.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 3.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 2.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 3.8 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 3.2 | GO:0043647 | inositol phosphate metabolic process(GO:0043647) |
| 0.0 | 0.8 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 25.0 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.3 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 1.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 2.4 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 1.6 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.5 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 3.7 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.7 | GO:0060674 | placenta blood vessel development(GO:0060674) |
| 0.0 | 3.8 | GO:0043279 | response to alkaloid(GO:0043279) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 2.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.2 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 4.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.6 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 8.0 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.7 | GO:0070671 | interleukin-12-mediated signaling pathway(GO:0035722) response to interleukin-12(GO:0070671) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.6 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.4 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.0 | 22.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 1.7 | 13.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.6 | 4.7 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.4 | 18.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 1.0 | 5.0 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.9 | 5.6 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.9 | 4.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.7 | 17.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.7 | 3.7 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.7 | 5.8 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 7.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.5 | 3.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.5 | 7.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 3.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.4 | 8.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.4 | 2.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 5.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.3 | 3.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 9.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 3.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 4.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.2 | 0.7 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.2 | 14.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 13.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 3.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 3.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 7.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 1.5 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 3.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 13.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 3.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 10.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 28.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 0.5 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.2 | 0.6 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.9 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 8.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 9.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 3.0 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 1.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 3.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 7.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 8.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 1.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 8.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 9.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 6.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 6.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 5.6 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 21.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 12.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.1 | 1.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 5.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 8.7 | GO:1904813 | ficolin-1-rich granule(GO:0101002) ficolin-1-rich granule lumen(GO:1904813) |
| 0.0 | 4.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 10.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 5.7 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 3.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 6.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 8.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 8.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.3 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 7.3 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 27.4 | GO:0005654 | nucleoplasm(GO:0005654) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 3.6 | 10.9 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 3.0 | 11.8 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.0 | 6.0 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 1.5 | 6.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 1.4 | 1.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.2 | 7.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.0 | 21.0 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.9 | 19.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.8 | 2.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.7 | 3.5 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.7 | 4.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.6 | 3.2 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.6 | 8.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.5 | 2.2 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.5 | 5.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.5 | 3.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.5 | 18.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.5 | 2.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.5 | 8.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.5 | 13.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 1.3 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.4 | 5.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 4.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.4 | 8.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.4 | 5.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.4 | 1.5 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.4 | 15.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 3.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 1.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.3 | 10.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.3 | 3.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.2 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 1.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 1.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.3 | 0.8 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.3 | 10.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.3 | 4.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 2.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 5.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 4.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.2 | 2.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 10.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 1.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 21.7 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.2 | 20.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.2 | 3.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 5.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 3.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 1.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 4.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 13.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 1.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 3.0 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 2.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 6.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 7.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 4.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 3.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 8.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 4.8 | GO:0061650 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 3.3 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 1.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 5.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 3.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 7.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 2.5 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 3.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 4.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 8.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 7.4 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 1.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 13.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 7.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 15.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 3.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 9.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.8 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 5.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 11.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 6.2 | GO:0060089 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 1.1 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 10.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 16.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 4.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 6.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.3 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 15.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.7 | 17.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.4 | 25.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 8.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 19.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.2 | 23.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 2.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 17.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.2 | 11.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.2 | 1.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 9.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 6.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 5.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 8.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 13.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 8.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 3.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 4.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 3.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 3.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.5 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 4.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 0.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 3.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.7 | 22.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.5 | 10.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.4 | 11.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 7.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 9.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.4 | 20.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 15.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.3 | 2.8 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 6.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 5.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 3.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.2 | 16.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.2 | 3.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 3.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 3.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 4.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 2.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 3.0 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 4.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 5.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 14.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 7.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 5.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 7.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 3.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 5.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 9.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 5.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 3.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 2.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 5.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |