Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
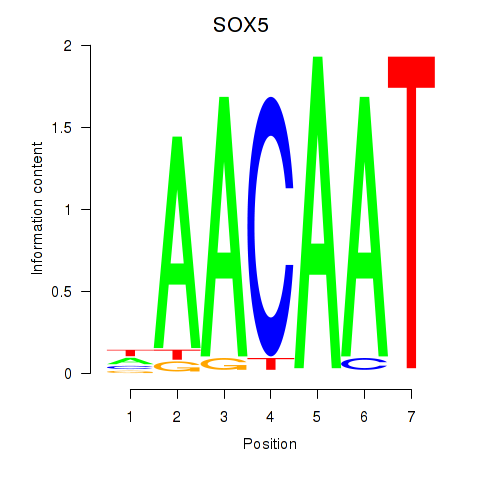
Results for SOX5
Z-value: 0.86
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg19_v2_chr12_-_23737534_23737550, hg19_v2_chr12_-_24103954_24103972 | 0.40 | 6.2e-10 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_6233828 | 15.92 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr14_-_92414055 | 15.84 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr14_-_92413727 | 14.81 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr7_+_111846741 | 12.97 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chrX_-_13835147 | 12.29 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr7_+_30960915 | 10.83 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr18_-_52989217 | 10.82 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr15_+_84115868 | 10.54 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr6_-_152957944 | 10.26 |
ENST00000423061.1
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr2_+_203499901 | 10.17 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr7_-_111846435 | 10.15 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr4_-_76598326 | 9.47 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_+_126240442 | 9.42 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_-_114790179 | 9.20 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chrY_+_15016013 | 9.20 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr6_-_99873145 | 9.14 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr3_-_18466026 | 9.02 |
ENST00000417717.2
|
SATB1
|
SATB homeobox 1 |
| chr10_-_13523073 | 8.27 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chrX_-_50557014 | 8.11 |
ENST00000376020.2
|
SHROOM4
|
shroom family member 4 |
| chr12_+_53443680 | 8.11 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_-_123339418 | 8.00 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr3_-_114343039 | 7.77 |
ENST00000481632.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr9_-_130712995 | 7.68 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr3_-_123339343 | 7.62 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr10_+_70480963 | 7.49 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chrX_-_50557302 | 7.45 |
ENST00000289292.7
|
SHROOM4
|
shroom family member 4 |
| chr12_+_53443963 | 7.40 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr14_-_92413353 | 7.31 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr18_-_53177984 | 7.30 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr4_-_2264015 | 6.88 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr18_+_42260861 | 6.79 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr2_+_30569506 | 6.78 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr11_+_46402482 | 6.72 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr19_+_18794470 | 6.68 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr1_-_156217822 | 6.62 |
ENST00000368270.1
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr2_-_157198860 | 6.62 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr1_-_12677714 | 6.60 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr16_-_11375179 | 6.56 |
ENST00000312511.3
|
PRM1
|
protamine 1 |
| chr17_-_10452929 | 6.52 |
ENST00000532183.2
ENST00000397183.2 ENST00000420805.1 |
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult |
| chrX_+_135251783 | 6.50 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr1_-_156217829 | 6.44 |
ENST00000356983.2
ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr1_-_156217875 | 6.43 |
ENST00000292291.5
|
PAQR6
|
progestin and adipoQ receptor family member VI |
| chr12_+_32655048 | 6.41 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr3_+_189507460 | 6.34 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr8_-_33424636 | 6.32 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chr6_-_32557610 | 6.24 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr11_+_131781290 | 6.24 |
ENST00000425719.2
ENST00000374784.1 |
NTM
|
neurotrimin |
| chr10_-_62149433 | 6.19 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_-_99594948 | 6.07 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_+_52445191 | 6.05 |
ENST00000243050.1
ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_+_31085714 | 6.01 |
ENST00000300850.5
ENST00000564189.1 ENST00000428260.1 |
ZNF646
|
zinc finger protein 646 |
| chr16_-_31085514 | 5.89 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chr8_+_85095769 | 5.85 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr14_+_61654271 | 5.62 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr1_+_9005917 | 5.60 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr14_+_22977587 | 5.56 |
ENST00000390504.1
|
TRAJ33
|
T cell receptor alpha joining 33 |
| chrX_+_135252050 | 5.50 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr12_-_11508520 | 5.50 |
ENST00000545626.1
ENST00000500254.2 |
PRB1
|
proline-rich protein BstNI subfamily 1 |
| chr2_+_27070964 | 5.46 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr4_-_186732048 | 5.24 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr18_-_52989525 | 5.22 |
ENST00000457482.3
|
TCF4
|
transcription factor 4 |
| chr20_-_3762087 | 5.20 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr4_+_165675269 | 5.19 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr20_+_17207665 | 5.15 |
ENST00000536609.1
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr4_-_87281196 | 5.15 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr8_+_85095497 | 5.09 |
ENST00000522455.1
ENST00000521695.1 |
RALYL
|
RALY RNA binding protein-like |
| chr6_+_89790490 | 5.04 |
ENST00000336032.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr15_-_86338134 | 4.99 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr11_+_126225529 | 4.97 |
ENST00000227495.6
ENST00000444328.2 ENST00000356132.4 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr6_+_89790459 | 4.96 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chrX_-_17878827 | 4.94 |
ENST00000360011.1
|
RAI2
|
retinoic acid induced 2 |
| chr2_-_208989225 | 4.92 |
ENST00000264376.4
|
CRYGD
|
crystallin, gamma D |
| chr17_+_33448593 | 4.85 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr1_+_15272271 | 4.84 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr1_+_11751748 | 4.77 |
ENST00000294485.5
|
DRAXIN
|
dorsal inhibitory axon guidance protein |
| chr20_+_17207636 | 4.72 |
ENST00000262545.2
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2 |
| chr3_+_181429704 | 4.59 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr1_-_226926864 | 4.56 |
ENST00000429204.1
ENST00000366784.1 |
ITPKB
|
inositol-trisphosphate 3-kinase B |
| chr8_+_85095553 | 4.50 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr1_+_156589051 | 4.39 |
ENST00000255039.1
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2 |
| chrX_-_106959631 | 4.36 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr15_-_86338100 | 4.29 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr14_-_23904861 | 4.27 |
ENST00000355349.3
|
MYH7
|
myosin, heavy chain 7, cardiac muscle, beta |
| chr14_+_37126765 | 4.23 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr3_-_99595037 | 4.22 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr22_+_43808014 | 4.17 |
ENST00000334209.5
ENST00000443721.1 ENST00000414469.2 ENST00000439548.1 |
MPPED1
|
metallophosphoesterase domain containing 1 |
| chr12_-_111358372 | 4.16 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr12_-_102874416 | 4.16 |
ENST00000392904.1
ENST00000337514.6 |
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr2_-_26205340 | 4.11 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr6_+_126070726 | 4.06 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr4_-_87281224 | 4.03 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr10_+_35415719 | 3.98 |
ENST00000474362.1
ENST00000374721.3 |
CREM
|
cAMP responsive element modulator |
| chr2_-_160472952 | 3.93 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr15_-_34610962 | 3.89 |
ENST00000290209.5
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr5_-_146461027 | 3.85 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr12_-_102874378 | 3.85 |
ENST00000456098.1
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr2_-_160473114 | 3.76 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr10_+_49514698 | 3.76 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr2_+_166428839 | 3.74 |
ENST00000342316.4
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_-_62332357 | 3.72 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr12_-_102872317 | 3.71 |
ENST00000424202.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chrX_+_135251835 | 3.66 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr1_-_151119087 | 3.62 |
ENST00000341697.3
ENST00000368914.3 |
SEMA6C
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr10_+_63661053 | 3.59 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr11_-_115375107 | 3.56 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr1_+_164528866 | 3.52 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_+_113970772 | 3.51 |
ENST00000504454.1
ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2
|
ankyrin 2, neuronal |
| chr10_-_116444371 | 3.47 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr4_+_71588372 | 3.46 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr11_-_82708519 | 3.41 |
ENST00000534301.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr8_-_93115445 | 3.36 |
ENST00000523629.1
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_-_46048116 | 3.33 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr11_+_112832202 | 3.32 |
ENST00000534015.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr12_-_52715179 | 3.30 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr4_+_25657444 | 3.28 |
ENST00000504570.1
ENST00000382051.3 |
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr8_-_72274467 | 3.26 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr7_+_29519662 | 3.25 |
ENST00000424025.2
ENST00000439711.2 ENST00000421775.2 |
CHN2
|
chimerin 2 |
| chr13_-_28545276 | 3.10 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr10_+_95517660 | 3.04 |
ENST00000371413.3
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr1_+_163038565 | 3.00 |
ENST00000421743.2
|
RGS4
|
regulator of G-protein signaling 4 |
| chr4_+_88754069 | 2.98 |
ENST00000395102.4
ENST00000497649.2 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr1_+_50574585 | 2.98 |
ENST00000371824.1
ENST00000371823.4 |
ELAVL4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_+_12949251 | 2.97 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr2_-_70475730 | 2.94 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chrX_+_107288239 | 2.93 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_28177255 | 2.92 |
ENST00000601459.1
|
AL109927.1
|
HCG2032222; PRO2047; Uncharacterized protein |
| chr17_-_39743139 | 2.92 |
ENST00000167586.6
|
KRT14
|
keratin 14 |
| chr11_+_46402583 | 2.87 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr19_-_51472823 | 2.86 |
ENST00000310157.2
|
KLK6
|
kallikrein-related peptidase 6 |
| chr12_-_8815404 | 2.79 |
ENST00000359478.2
ENST00000396549.2 |
MFAP5
|
microfibrillar associated protein 5 |
| chr5_-_134871639 | 2.79 |
ENST00000314744.4
|
NEUROG1
|
neurogenin 1 |
| chr10_+_35415978 | 2.75 |
ENST00000429130.3
ENST00000469949.2 ENST00000460270.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_56954867 | 2.68 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr11_+_46402744 | 2.65 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_+_112832090 | 2.62 |
ENST00000533760.1
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr12_+_52306113 | 2.59 |
ENST00000547400.1
ENST00000550683.1 ENST00000419526.2 |
ACVRL1
|
activin A receptor type II-like 1 |
| chr5_+_67588391 | 2.54 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr17_+_30814707 | 2.51 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chrX_+_107683096 | 2.50 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr2_+_33661382 | 2.45 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr4_+_41614909 | 2.45 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_-_70475701 | 2.44 |
ENST00000282574.4
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chrX_-_53711064 | 2.44 |
ENST00000342160.3
ENST00000446750.1 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr15_+_66994663 | 2.42 |
ENST00000457357.2
|
SMAD6
|
SMAD family member 6 |
| chr1_-_40367530 | 2.41 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr3_+_89156799 | 2.36 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr4_+_88754113 | 2.34 |
ENST00000560249.1
ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr9_-_37465396 | 2.32 |
ENST00000307750.4
|
ZBTB5
|
zinc finger and BTB domain containing 5 |
| chr4_-_68749745 | 2.30 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr12_-_71031185 | 2.25 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr1_-_167883353 | 2.24 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr11_+_128563948 | 2.21 |
ENST00000534087.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_29519486 | 2.20 |
ENST00000409041.4
|
CHN2
|
chimerin 2 |
| chr17_+_3118915 | 2.19 |
ENST00000304094.1
|
OR1A1
|
olfactory receptor, family 1, subfamily A, member 1 |
| chrX_+_107288197 | 2.17 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr9_-_123476719 | 2.17 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr3_+_189507523 | 2.14 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr11_-_35440796 | 2.10 |
ENST00000278379.3
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_+_128563652 | 2.09 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_39408470 | 2.08 |
ENST00000257408.4
|
KLB
|
klotho beta |
| chr9_-_72287191 | 2.07 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr9_+_82188077 | 2.07 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr8_+_103563792 | 2.05 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr16_+_2083265 | 2.05 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr3_+_171561127 | 2.00 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr12_-_102591604 | 2.00 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr20_-_18477862 | 1.98 |
ENST00000337227.4
|
RBBP9
|
retinoblastoma binding protein 9 |
| chr9_-_123476612 | 1.97 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr1_-_167883327 | 1.95 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr22_-_40859415 | 1.94 |
ENST00000402630.1
ENST00000407029.1 |
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr9_+_27109133 | 1.90 |
ENST00000519097.1
ENST00000380036.4 |
TEK
|
TEK tyrosine kinase, endothelial |
| chr10_+_95517616 | 1.88 |
ENST00000371418.4
|
LGI1
|
leucine-rich, glioma inactivated 1 |
| chr9_-_124989804 | 1.86 |
ENST00000373755.2
ENST00000373754.2 |
LHX6
|
LIM homeobox 6 |
| chr13_-_74708372 | 1.85 |
ENST00000377666.4
|
KLF12
|
Kruppel-like factor 12 |
| chr1_+_10271674 | 1.80 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr16_+_1832902 | 1.77 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr2_-_2334888 | 1.77 |
ENST00000428368.2
ENST00000399161.2 |
MYT1L
|
myelin transcription factor 1-like |
| chr20_-_45985172 | 1.74 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chrX_+_24483338 | 1.71 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr10_+_35416223 | 1.68 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr6_+_122720681 | 1.64 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr12_-_71031220 | 1.62 |
ENST00000334414.6
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chrX_-_112084043 | 1.61 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr11_-_35441597 | 1.53 |
ENST00000395753.1
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr4_-_68749699 | 1.53 |
ENST00000545541.1
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr13_+_73629107 | 1.51 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr15_+_66994561 | 1.50 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr1_+_209757051 | 1.49 |
ENST00000009105.1
ENST00000423146.1 ENST00000361322.2 |
CAMK1G
|
calcium/calmodulin-dependent protein kinase IG |
| chr7_-_131241361 | 1.49 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr12_-_51477333 | 1.48 |
ENST00000228515.1
ENST00000548206.1 ENST00000546935.1 ENST00000548981.1 |
CSRNP2
|
cysteine-serine-rich nuclear protein 2 |
| chr10_+_35416090 | 1.46 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr3_+_189349162 | 1.46 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr3_+_89156674 | 1.41 |
ENST00000336596.2
|
EPHA3
|
EPH receptor A3 |
| chr12_-_111926342 | 1.41 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr7_-_11871815 | 1.41 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr8_-_72274095 | 1.36 |
ENST00000303824.7
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr4_-_101439242 | 1.32 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr8_-_30706608 | 1.32 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr3_+_134514093 | 1.29 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr12_-_91451758 | 1.28 |
ENST00000266719.3
|
KERA
|
keratocan |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 3.2 | 38.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 2.5 | 9.9 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 2.5 | 9.9 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 2.3 | 11.5 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 2.2 | 6.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 2.2 | 6.6 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 2.0 | 12.2 | GO:0030421 | defecation(GO:0030421) |
| 2.0 | 10.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 2.0 | 9.9 | GO:0030070 | insulin processing(GO:0030070) |
| 2.0 | 15.6 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 1.6 | 12.9 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.4 | 4.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 1.3 | 3.8 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 1.2 | 10.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.2 | 6.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 1.2 | 12.3 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.2 | 3.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 4.3 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 1.0 | 10.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.9 | 2.8 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.9 | 3.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.9 | 4.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.9 | 6.1 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.8 | 2.5 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.8 | 3.9 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.8 | 3.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.8 | 4.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.7 | 5.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.6 | 1.8 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.6 | 4.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.6 | 9.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.6 | 4.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.6 | 9.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 | 4.4 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.5 | 6.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.5 | 4.6 | GO:0072513 | semicircular canal morphogenesis(GO:0048752) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.5 | 3.9 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.5 | 2.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 6.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 3.9 | GO:0003183 | mitral valve morphogenesis(GO:0003183) |
| 0.4 | 2.6 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.4 | 1.3 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.4 | 9.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 3.0 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 3.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.3 | 3.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.3 | 2.7 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.3 | 2.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 4.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.3 | 2.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.3 | 4.6 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.3 | 15.7 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.3 | 1.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 5.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.3 | 0.8 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 1.7 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.2 | 5.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 2.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 4.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 19.5 | GO:0032760 | positive regulation of tumor necrosis factor production(GO:0032760) |
| 0.2 | 1.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 7.6 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.2 | 1.9 | GO:0021853 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.2 | 2.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 5.0 | GO:0097503 | sialylation(GO:0097503) |
| 0.2 | 6.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.2 | 2.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 3.9 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 2.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 5.4 | GO:1904037 | positive regulation of epithelial cell apoptotic process(GO:1904037) |
| 0.2 | 5.8 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.2 | 0.5 | GO:0003264 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.2 | 0.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 2.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 3.7 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 1.8 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 3.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 4.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 2.9 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 1.0 | GO:0048505 | positive regulation of activin receptor signaling pathway(GO:0032927) regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.1 | 4.9 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 1.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 10.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 3.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 2.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 4.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 3.6 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 1.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 5.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 11.7 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.1 | 0.7 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 3.0 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.5 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 2.9 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 18.0 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.1 | 14.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.1 | 4.8 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 3.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 2.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 1.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 13.9 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 2.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 5.0 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 4.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 4.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 3.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 1.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 2.0 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.8 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.2 | GO:0072608 | interleukin-10 secretion(GO:0072608) regulation of interleukin-10 secretion(GO:2001179) negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.1 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 1.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.5 | GO:0042127 | regulation of cell proliferation(GO:0042127) |
| 0.0 | 1.1 | GO:0042476 | odontogenesis(GO:0042476) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 38.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 3.6 | 10.8 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 2.2 | 6.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.6 | 11.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 1.1 | 13.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.8 | 10.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.8 | 5.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.6 | 2.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.6 | 27.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.6 | 2.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 6.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.3 | 10.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 6.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 2.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 2.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.3 | 4.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 7.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.2 | 15.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 9.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 2.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 4.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.2 | 1.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 3.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 4.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 32.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.2 | 3.5 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 4.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 9.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 3.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.1 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 10.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 5.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 12.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 16.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 3.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 7.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 8.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.6 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 0.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 5.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 5.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 4.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 3.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 12.2 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 3.1 | 15.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 2.2 | 12.9 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.8 | 23.3 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.2 | 5.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 1.1 | 5.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 1.1 | 16.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 1.1 | 5.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 1.0 | 4.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.9 | 6.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.8 | 2.5 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.8 | 3.9 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.8 | 9.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.7 | 4.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 3.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.6 | 2.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 2.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 2.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.5 | 15.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.5 | 2.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 4.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.4 | 1.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.4 | 3.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.4 | 10.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 3.9 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 14.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.3 | 5.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 5.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 13.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 3.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 6.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 10.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.3 | 1.6 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.3 | 1.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 16.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.3 | 0.8 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.2 | 4.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 5.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 8.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 38.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.6 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 6.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 4.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 19.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 10.2 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.2 | 4.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 4.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 3.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 15.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.8 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 5.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 5.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 3.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 6.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.2 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 13.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 9.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 3.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 3.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 3.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 10.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 2.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.1 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 5.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.9 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 3.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 4.6 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.4 | 22.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 73.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.3 | 4.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 10.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 5.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 12.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 14.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.2 | 25.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 16.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 11.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 6.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 6.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 9.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 10.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 5.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 3.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 3.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 3.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.8 | 10.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.6 | 11.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.5 | 22.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.4 | 9.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 5.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.3 | 7.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 9.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 6.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.2 | 14.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 4.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 3.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 8.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 9.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 6.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 11.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 4.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 6.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 5.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 3.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |