Project
GNF SymAtlas + NCI-60 cancer cell lines, comparison of cancers vs non-cancers, human (Su, 2004; Ross, 2000)
Navigation
Downloads
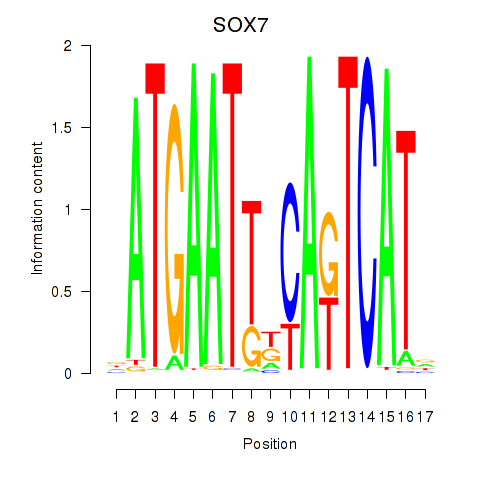
Results for SOX7
Z-value: 1.07
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg19_v2_chr8_-_10588010_10588030 | 0.69 | 5.5e-32 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_66951474 | 17.47 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr21_+_34144411 | 14.58 |
ENST00000382375.4
ENST00000453404.1 ENST00000382378.1 ENST00000477513.1 |
C21orf49
|
chromosome 21 open reading frame 49 |
| chr18_+_32820990 | 8.44 |
ENST00000601719.1
ENST00000591206.1 ENST00000330501.7 ENST00000261333.6 ENST00000355632.4 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr8_-_82359662 | 8.00 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chr16_-_20364122 | 7.83 |
ENST00000396138.4
ENST00000577168.1 |
UMOD
|
uromodulin |
| chr11_-_62474803 | 7.34 |
ENST00000533982.1
ENST00000360796.5 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr22_+_23101182 | 7.04 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr16_+_33020496 | 6.88 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr10_+_5005445 | 6.87 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr16_-_20364030 | 6.30 |
ENST00000396134.2
ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD
|
uromodulin |
| chr1_+_156698708 | 5.89 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr1_+_100598691 | 5.72 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr10_-_71169031 | 5.69 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr15_+_25068773 | 5.69 |
ENST00000400100.1
ENST00000400098.1 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr16_-_279405 | 5.63 |
ENST00000430864.1
ENST00000293872.8 ENST00000337351.4 ENST00000397783.1 |
LUC7L
|
LUC7-like (S. cerevisiae) |
| chr11_+_3011093 | 5.57 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr5_+_175511859 | 5.57 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
| chr1_+_158901329 | 5.41 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr9_+_105757590 | 5.39 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr16_-_33647696 | 5.32 |
ENST00000558425.1
ENST00000569103.2 |
RP11-812E19.9
|
Uncharacterized protein |
| chr1_+_11724167 | 4.81 |
ENST00000376753.4
|
FBXO6
|
F-box protein 6 |
| chr10_-_5046042 | 4.69 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr8_-_102803163 | 4.19 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr8_-_19459993 | 4.18 |
ENST00000454498.2
ENST00000520003.1 |
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr6_+_150690133 | 3.79 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr5_+_161494521 | 3.62 |
ENST00000356592.3
|
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr4_-_47983519 | 3.48 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr6_+_150690028 | 3.37 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr10_+_96162242 | 3.31 |
ENST00000225235.4
|
TBC1D12
|
TBC1 domain family, member 12 |
| chr6_+_29555683 | 3.21 |
ENST00000383640.2
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2 |
| chr6_+_131894284 | 3.12 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr3_-_123411191 | 3.00 |
ENST00000354792.5
ENST00000508240.1 |
MYLK
|
myosin light chain kinase |
| chr8_+_12803176 | 2.81 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr5_+_161494770 | 2.77 |
ENST00000414552.2
ENST00000361925.4 |
GABRG2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr7_-_143966381 | 2.69 |
ENST00000487179.1
|
CTAGE8
|
CTAGE family, member 8 |
| chr2_+_3705785 | 2.65 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr19_-_2783363 | 2.50 |
ENST00000221566.2
|
SGTA
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr14_+_50291993 | 2.49 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr1_+_10509971 | 2.23 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr2_-_166930131 | 2.11 |
ENST00000303395.4
ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit |
| chr4_+_76995855 | 1.88 |
ENST00000355810.4
ENST00000349321.3 |
ART3
|
ADP-ribosyltransferase 3 |
| chr6_-_46048116 | 1.65 |
ENST00000185206.6
|
CLIC5
|
chloride intracellular channel 5 |
| chr13_-_37633567 | 1.17 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr19_-_22379753 | 1.09 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr17_-_39623681 | 0.79 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chrY_+_14813160 | 0.70 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr8_+_77593448 | 0.64 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr10_-_17243579 | 0.59 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr12_-_58027138 | 0.56 |
ENST00000341156.4
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chr14_-_106573756 | 0.54 |
ENST00000390601.2
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chrX_+_69353284 | 0.20 |
ENST00000342206.6
ENST00000356413.4 |
IGBP1
|
immunoglobulin (CD79A) binding protein 1 |
| chr15_+_59439899 | 0.13 |
ENST00000599727.1
|
C15ORF31
|
C15ORF31 |
| chr11_+_7618413 | 0.12 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 14.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 1.9 | 5.7 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 1.7 | 11.6 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 1.6 | 17.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 1.0 | 4.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.9 | 2.6 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.8 | 2.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.6 | 3.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.6 | 7.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.4 | 5.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.4 | 3.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.3 | 9.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.3 | 6.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 7.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 4.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.2 | 2.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 2.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 5.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 3.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 6.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 3.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.1 | 7.6 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 3.2 | GO:0007618 | mating(GO:0007618) |
| 0.0 | 1.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 4.2 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 5.7 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 4.9 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.5 | 11.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.3 | 4.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 3.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 6.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 14.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 6.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.1 | GO:0043194 | voltage-gated sodium channel complex(GO:0001518) axon initial segment(GO:0043194) |
| 0.1 | 1.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 7.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 8.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.1 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 17.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.3 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 5.4 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 17.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 1.9 | 11.6 | GO:0047115 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 1.4 | 4.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 1.2 | 7.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 1.2 | 5.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.1 | 5.7 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.8 | 2.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.8 | 5.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.8 | 14.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.6 | 3.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 2.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 6.4 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.4 | 5.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.4 | 1.9 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.3 | 6.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.3 | 3.5 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 8.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 6.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 4.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 2.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 3.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 7.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 5.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 4.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 17.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.3 | 7.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 6.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 3.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 5.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.1 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |